We revealed that binding of agitoxin-2 to a K+ channel uses an induced-fit pathway by single-molecule analysis via high-speed AFM.
Abstract
Agitoxin-2 (AgTx2) from scorpion venom is a potent blocker of K+ channels. The docking model has been elucidated, but it remains unclear whether binding dynamics are described by a two-state model (AgTx2-bound and AgTx2-unbound) or a more complicated mechanism, such as induced fit or conformational selection. Here, we observed the binding dynamics of AgTx2 to the KcsA channel using high-speed atomic force microscopy. From images of repeated binding and dissociation of AgTx2 to the channel, single-molecule kinetic analyses revealed that the affinity of the channel for AgTx2 increased during persistent binding and decreased during persistent dissociation. We propose a four-state model, including high- and low-affinity states of the channel, with relevant rate constants. An induced-fit pathway was dominant and accelerated binding by 400 times. This is the first analytical imaging of scorpion toxin binding in real time, which is applicable to various biological dynamics including channel ligands, DNA-modifier proteins, and antigen-antibody complexes.
INTRODUCTION
Ion channels regulate membrane potential by mediating the permeation of specific ion species via their transmembrane pore with gating. Gating behavior is regulated by a range of factors, including membrane voltage, membrane tension, and the binding of ligands (1, 2). The inappropriate permeation of ions (due to errors in the gating of these channels) can lead to dysregulation of the membrane potential, resulting in critical dysfunction of cells (e.g., Long-QT syndrome, Bartter syndrome, and periodic paralysis) (3).
Channel blockers are important ligands that hamper ion channel function. Scorpion venom is a cocktail of many ion channel blockers (4–6), and approximately 250 ligands with affinities to K+ channels have been identified across several species of scorpions (7–9). Agitoxins (AgTx) are peptides found in Leiurus quinquestriatus var. hebraeus venom, which act as potent and selective blockers of the Shaker-type voltage-gated Kv1.3 and Kv1.1 channels (10). Agitoxin-2 (AgTx2; 38 amino acid residues; Fig. 1A) is a most studied AgTx that binds to the extracellular vestibule of the Shaker-related K+ channels through a combination of electrostatic, hydrogen bonding, and hydrophobic interactions (11, 12) and blocks ion permeation through the channel, causing many physiological dysfunctions.
Fig. 1. Schematic illustrations and AFM imaging of AgTx2 binding to the extracellular surface of the KcsA channel.

(A) X-ray crystallographic structure of the KcsA channel [Protein Data Bank (PDB) ID: 1K4C] (14) and solution structure of AgTx2 obtained by NMR (1AGT) (15). Gray dots are K+ ions. Only two diagonal subunits of the tetrameric channel are shown. The side chain of the K27 residue on AgTx2 is shown as a stick structure. (B) Arrangements of the reconstituted channel with (right) or without (left) AgTx2 on the substrate. (C) Typical AFM images of the channel with (right) or without (left) AgTx2. The KcsA channel is reconstituted in the DMPC bilayer. The AgTx2 is added to an imaging buffer. Height profiles along the white dotted lines in the AFM images are shown below the images. The background illustration behind the height profiles indicates the corresponding structures of the channel and AgTx2. The binding model (model II) (12) was downloaded from B. Roux’s web page (http://thallium.bsd.uchicago.edu/RouxLab/sub/gallery/agtx_shaker.html). Imaged samples were in 10 mM Hepes (pH 7.5) containing 200 mM KCl and 20 nM AgTx2. Scale bars, 2 nm. Z scale, 15 Å.
The KcsA channel is a prototypical bacterial channel and a Shaker-related K+ channel as well as Kv1.1 and Kv1.3 (13, 14). Purification of the recombinant KcsA channel is easier than Kv1.1 and Kv1.3 itself, and thus, KcsA is widely used as a model of the Shaker-related K+ channel. A high-resolution crystallographic structure of the KcsA is available (14), facilitating understanding of the structure-function relationship of K+ channels. The KcsA channel has been used to mimic the AgTx2-binding site of the Shaker K+ channel and its mammalian homolog by introducing triple mutations (Q58A, T61S, and R64D) (11).
Nuclear magnetic resonance (NMR) spectroscopy has revealed the solution structure of AgTx2 (15, 16). Eriksson and Roux (12) generated docking structures of AgTx2 and a modeled Shaker K+ channel using the structure of AgTx2. The authors reported that the F25 side chain of AgTx2 often changes its orientation angle relative to the body of the AgTx2 molecule in the docked structures and claimed that this structural change in the toxin suggests the presence of an “induced-fit” mechanism. The induced-fit mechanism is an important concept used to describe ligands binding to biological molecules. The interaction between AgTx2 molecules and an external channel surface provides an intriguing opportunity to examine whether the concept of the induced-fit mechanism is applicable. In the induced-fit model, a receptor molecule undergoes conformational changes after ligand binding, leading to a higher affinity for the ligand. Eriksson and Roux measured changes in the AgTx2 structure (AgTx2 was near the channel for ~3 ns), but the simulation time was not sufficient to detect conformational changes of the relevant part of the channel after AgTx2 binding. Thus, the proposed induced-fit mechanism upon AgTx2 binding to potassium channels remains elusive. Besides the induced fit, conformational selection has been proposed as a complicated binding mechanism. With conformational selection, the receptor molecule undergoes transitions between two conformational states with high and low ligand affinity, even in the absence of ligands, and the ligand preferentially binds to the high-affinity receptor. In contrast to simple two-state dynamics of ligand binding, the induced-fit and conformational selection mechanisms indicate advanced binding dynamics of biological molecules (17–24) and provide opportunities for fine-tuning ligand binding in various physiological functions. Thus, the molecular mechanisms underlying these concepts need to be examined by dynamic experimental and analytical methods.
Basically, both the conformational selection and induced-fit mechanisms can be described by a four-state model (17), in which the receptor has two distinct conformational states, having high and low ligand affinity (24). To investigate such binding dynamics, including binding to the receptor and dissociation into aqueous solution, a large amount of statistics from long-duration measurements are needed. Distinguishing such dynamics using conventional macroscopic measurements, such as biochemical binding assays, is technically difficult. For example, identifying transient states, such as a low-affinity binding state during the induced-fit pathway, might be a key to understanding the dynamics of binding; however, such a transient state is easily hidden in a large ensemble of other states and often exists as a small fraction in the whole system because the macroscopic measurements are unsynchronized. To identify whether the transient state is a ligand-bound state, an understanding of the binding dynamics, including the transient state at the nanoscopic or single-molecule level, is critically important.
To capture the microscopic dynamics of binding, single-molecule observation is an attractive technique for investigating not only the macroscopic affinity, measured as the dissociation constant (Kd), but also the rate constants (kass and kdiss) at the single-molecule level. However, even with single-channel current recordings (25), distinguishing the dynamics of channel gating from the binding of a channel blocker needs elaborated analyses. Fluorescence measurements are also used for studying single-molecule binding dynamics (26–29), but molecules of interest must be labeled with fluorophores that might disrupt the intrinsic interactions between those molecules and the channel. By contrast, high-speed atomic force microscopy (HS-AFM) can image the surface structure of nonlabeled biomolecules in a solution with high spatiotemporal resolution (horizontal, ~1 nm; vertical, 0.1 nm; temporal, ~10 fps) (30, 31). Therefore, various biological events have been successfully imaged by HS-AFM (32), e.g., structural changes in soluble and membrane proteins (33–38) and binding dynamics of soluble proteins (39, 40). Furthermore, HS-AFM imaging can be used to analyze binding dynamics of ligands even if channels are fully closed and enables us to investigate cooperativity among channels. These are some of the advantages of using HS-AFM over single-channel current recordings. Consequently, HS-AFM represents a powerful tool for studying channel-ligand interactions.
In this study, we observed binding dynamics of AgTx2 to a K+ channel using HS-AFM and examined the single-molecule dynamics using a recently developed analytical method. Visualization of discrete ligand binding and unbinding events using HS-AFM allowed detection of hidden underlying processes of the channel after toxin binding or unbinding. We conclude that the induced-fit pathway contributes to the efficient AgTx2 binding to the KcsA channel, and the structural relevance of the induced-fit mechanism is discussed.
RESULTS
HS-AFM captures the single-molecule dynamics of the binding of AgTx2 to the K+ channel
AgTx2 binds to the extracellular surface of the channel (Fig. 1A) through electrostatic and hydrophobic interactions (12, 16, 41). To observe the dynamic AgTx2 binding on the KcsA channel by HS-AFM, we reconstituted the KcsA channel into a 1,2-dimyristoyl-sn-glycero-3-phosphocholine (DMPC) bilayer on a mica surface with its toxin-binding surface upward using a previously developed method (41). To suppress lateral diffusion of the channel in the membrane, we attached channels with a His-tag on the mica substrate via Ni2+–His-tag interactions (Fig. 1B). Since AgTx2 was added to the imaging buffer, it could bind to and dissociate from the channel during HS-AFM imaging. We supposed that the binding of AgTx2 to the channel elevates the apparent height of the channel by ~1.0 nm, as illustrated in Fig. 1B, because of the diameter of AgTx2.
Typical HS-AFM images of the reconstituted KcsA channels, in the presence of AgTx2 in buffer solution, showed two different features (Fig. 1C). The KcsA channel forms a homotetrameric oligomer (1), and the AFM images, while AgTx2 dissociates, showed four particles aligned as a square (Fig. 1C, top left). From the height profile of the AFM image along the extracellular channel surface (Fig. 1C, bottom), the distance between the tops of two diagonal subunits was 3.6 nm, which is in good agreement with the x-ray crystallographic structure of the channel (1K4C, the distance between two diagonal P55 is 3.6 nm) (14). The extracellular vestibule was concave ~0.5 nm from the top of the subunits. When AgTx2 bound to the extracellular vestibule of the channel, the channel-ligand complex surface appeared to be convex (Fig. 1C, top right) and 1.3 nm higher at the center of the channel than in the unbound state (Fig. 1C, bottom).
To analyze the binding dynamics quantitatively, we collected AFM movies showing repeated transitions of the AgTx2-unbound to AgTx2-bound state and its reverse. During continuous imaging in AgTx2-containing buffer, the channels repeatedly showed two distinct forms (Fig. 2A, top). One was a concave vestibule, and the other had a convex surface (white arrowheads), as described in the former section. To facilitate analysis, we transformed the binding transitions from a time series of two-dimensional (2D) images to simple 1D data. We collected time courses of averaged height around the center of the AgTx2-bound or AgTx2-unbound surface of the channels h(t) (Fig. 2A, bottom). The time course of h(t) showed binary transitions with a height difference of approximately 0.6 nm. [In the former section, we mentioned that the height change at the center of the channel upon AgTx2 binding was 1.3 nm (Fig. 1C). The smaller averaged height change upon AgTx2 binding (Fig. 2A) was due to the curved surface of the channel and AgTx2.] Thus, we concluded that these successive images and the distinct height changes demonstrated repeated AgTx2 binding to and dissociation from the channel. Other examples of binding transitions for 10 to 1000 nM AgTx2 are shown in fig. S1.
Fig. 2. HS-AFM captured AgTx2 binding to the channel with single-molecule resolution.
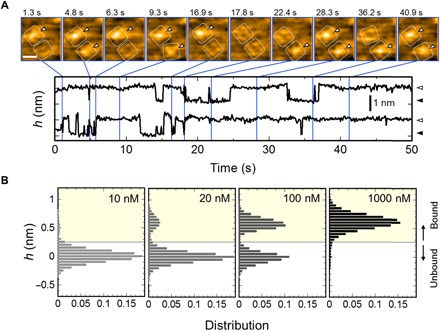
(A) Time-lapse images of AgTx2 binding to and dissociation from the KcsA channels (top; see also movie S1) and time courses of the averaged height h (nm) around the center of the extracellular surface (0.8 nm by 0.8 nm area) of two corresponding K+ channels (bottom). White dotted squares represent regions of interest for visualization of the tetrameric channels. Spontaneous AgTx2 bindings on the channels are indicated by white arrowheads (top). These two channels showed discrete changes in the h (nm), representing bound and unbound states of the channels (bottom). The bathing electrolyte solution contains 10 mM Hepes (pH 7.5) and 100 mM KCl. The scale bar in the first AFM image represents 5 nm. (B) Height histograms at 10, 20, 100, and 1000 nM AgTx2. The height of the extracellular surface of the AgTx2-unbound channel was set to 0 nm. Height transitions used in the histograms were measured in 10 mM Hepes (pH 7.5) containing 200 mM KCl. The number of data frames used for the histograms were 8604, 250,846, 33,701 and 35,453 for 10, 20, 100, and 1000 nM AgTx2, respectively.
We analyzed the AgTx2 concentration dependency of the state distributions using height transitions (Fig. 2B). At 10 nM AgTx2, the histogram showed a single distribution with a peak at 0 nm, whereas at 20 and 100 nM AgTx2, the distribution was bimodal with two peaks at 0 and 0.6 nm. At a higher concentration, 1000 nM AgTx2 produces a single distribution centered at 0.6 nm. Here, the AgTx2-bound and AgTx2-unbound states were defined as h > 0.25 and h ≤ 0.25 nm, respectively. The probability of the AgTx2-bound state of the channel (Pbound) was calculated as Pbound = Abound/(Abound + Aunbound), where Abound and Aunbound are the total time of the peaks above and below the threshold value (h = 0.25 nm), respectively. The Pbound values were 0.04, 0.16, 0.50, and 0.96 at AgTx2 concentrations of 10, 20, 100, and 1000 nM, respectively, indicating a Kd value of 100 nM. This Kd value is comparable to the previously reported Kd value using macroscopic measurements (11).
The K+ channel changes its affinity to AgTx2 after binding: A four-state binding model
To address whether the binding dynamics could be described by a simple two-state model comprising the AgTx2-bound and AgTx2-unbound states, we performed event-oriented analysis of the binding dynamics (42, 43). The time-course analysis of the height changes suggested two transitions: from AgTx2-bound to AgTx2-unbound and from AgTx2-unbound to AgTx2-bound. We refer to these transitions as “events.” In the event-oriented analysis, the height transition traces were synchronized at either the binding or the dissociation instance (event) (Fig. 3, B and C). Ensemble averaging of these traces reveals the time course of the binding probability, Pbound(Δt), as a function of time measured from the moment of the event when tpersist is 0 s (see below for the definition of tpersist) (42, 43).
Fig. 3. Event-oriented analysis of AgTx2 binding.
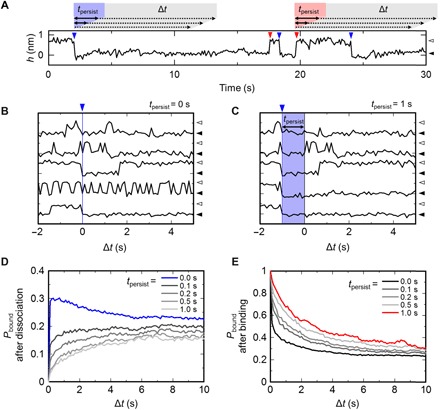
(A) Representative data of height transitions and definitions for the event-oriented analysis. The AgTx2-bound and AgTx2-unbound states are indicated by white and black arrowheads, respectively. Red and blue arrowheads indicate binding and dissociation events, respectively. For an arbitrary dissociation event (blue arrowhead), tpersist was defined from the instance of dissociation (blue). The time origin of Δt was the end of tpersist. The time variable definitions are the same as for the binding event (red arrowhead). (B and C) A representative dataset and the event-oriented analysis. The AgTx2-bound and AgTx2-unbound states are indicated by white and black arrowheads, respectively. All dissociation events were detected from the long-lasting height transition data, and strips of data before and after the event were collected and aligned at the dissociation events (Δt = 0) (blue arrowhead). Five examples are shown. For tpersist = 0 s (B), all the strips were ensemble-averaged, providing Pbound(Δt) for tpersist = 0 s. For tpersist = 1 s (C), strips of data persisting in the unbound state longer than tpersist after the dissociation event were used for ensemble averaging. (D) Time course of the binding probability after dissociation. Pbound(Δt) for different tpersist values ranging from immediately after dissociation (tpersist = 0 s) to 1 s from dissociation (tpersist = 1 s) are shown as a function of Δt measured from the end of tpersist. As tpersist increased, the time course of Pbound(Δt) could be expressed by a single-exponential function. (E) Time course of the binding probability after binding. Pbound(Δt) for different tpersist values from immediately after binding (tpersist = 0 s) to 1 s (tpersist = 1 s) after binding are shown. The binding transitions were measured in 10 mM Hepes (pH 7.5) containing 200 mM KCl and 20 nM AgTx2.
If the binding dynamics are described by a simple two-state model involving the AgTx2-bound and AgTx2-unbound states, Pbound exponentially increases from 0 to equilibrium Pbound (0.16 for 20 nM AgTx2 condition) with the passage of time from immediately after the dissociation (tpersist = 0 s). Pbound(Δt) did not follow a single-exponential curve (Fig. 3D, blue line). Pbound between 0 and 5 s after the dissociation event was greater than the equilibrium Pbound (0.16), indicating that the affinity of the channel for AgTx2 immediately after dissociation was greater than the equilibrium state of the channel. Thus, the channel-ligand system must have more than two states (i.e., more than only the AgTx2-bound and AgTx2-unbound states) in the system.
To analyze the complicated binding dynamics in greater detail, we introduce the time after the dissociation event that is spent in the unbound or bound state persistently and without transitions (persistent time or tpersist) (Fig. 3A). In the event-oriented analysis, the height transition traces remaining in the unbound state for longer than tpersist were collected and ensemble-averaged (Fig. 3, B and C), and the time course of Pbound was expressed as a function of time (Δt) measured from the end of tpersist (Fig. 3, D and E). This analysis reveals whether the affinity of the channel for AgTx2 changed during the persistent time that elapsed from the dissociation event. We assigned tpersist values of up to 1 s (Fig. 3A) and analyzed the dependence of Pbound(Δt) on variations of tpersist (Fig. 3D, gray lines). We found that if tpersist < 0.1 s, the probability of binding rapidly increased, meaning that the affinity of the channel for AgTx2 remained high after dissociation of AgTx2. By contrast, when tpersist = 0.5 s, the shape of Pbound(Δt) was almost the same as that of tpersist = 1.0 s, and Pbound(Δt) became an almost single-exponential curve if tpersist > 1 s (Fig. 3D, lightest gray line). These results suggest that the affinity of the channel decreases during persistent dissociation.
Similarly, we performed the event-oriented analysis immediately after the binding event (Fig. 3E). At tpersist < 0.1 s, Pbound very rapidly decreased from 1 upon binding, reaching an equilibrium Pbound over time. This finding demonstrates that the affinity of the channel was still low if AgTx2 bound to the channel for a short period. A longer tpersist resulted in a greater Pbound(Δt) (Fig. 3E, red line), indicating that the affinity of the channel to AgTx2 was greater during persistent binding. Thus, persistent binding of AgTx2 increased the affinity of the channel.
Overall, the event-oriented analysis showed that the affinity of the channel for AgTx2 increased during persistent binding and decreased during persistent dissociation, indicating that the binding model could not be described by the simple two-state (AgTx2-bound and AgTx2-unbound) model. Instead, to explain the binding dynamics, a plausible binding model should contain at least two conformational states of the channel with different affinities to AgTx2. In addition, there are AgTx2-bound and AgTx2-unbound states for each conformational state of the channel.
The event-oriented analysis strongly suggests that the K+ channel adopts at least four states with high and low affinities for AgTx2 (Fig. 4A). A four-state model involves two conformational states of the channel with high and low affinity to AgTx2 (CH and CL, respectively) and AgTx2-bound states for each conformation (CHTx and CLTx). Furthermore, we confirmed that both the bound and unbound states do not have more than two states by analyzing their dwell time distributions (fig. S2, A and B), indicating that the binding dynamics are well described by a four-state model. Therefore, we calculated the rate constants using a four-state model. The time courses of Pbound calculated by using the optimized rate constants are almost the same as the measured lines (fig. S2, C and D), indicating that the four-state model sufficiently reproduces the transition dynamics. To confirm whether the rate constants were reliable, we further calculated Pbound using the four-state model and compared it with the experimental Pbound values for AgTx2 in the AgTx2 concentration range from 10 to 1000 nM. As shown in Fig. 4B (black and white circles), the calculated Pbound values accurately reproduced the experimental Pbound. Furthermore, the binding transitions were also reproduced by the rate constants (Fig. 4C); the experimental and simulated binding transitions showed a similar pattern for various concentrations of AgTx2 (10, 20, 100, and 100 nM from the top to the bottom traces in Fig. 4C). Additional examples of binding transitions are shown in the Supplementary Materials (fig. S3). From these results, the four-state model and rate constants were consistent with the experimental data.
Fig. 4. The four-state binding model includes high- and low-affinity channels.

(A) The four-state model with high and low affinity of AgTx2 binding. The optimized rate constants are shown. (B) The binding probability Pbound evaluated by experimental and theoretical calculations using the four-state model. The experimental Pbound values were calculated from the height histograms in Fig. 2B. (C) Binding transitions obtained by HS-AFM imaging and simulation. The rate constants as shown in (A) were used for the simulation at 10, 20, 100, and 1000 nM AgTx2 (top to bottom). White and black arrowheads indicate the AgTx2-bound and AgTx2-unbound states of the channel, respectively. Red and blue points indicate the high- and low-affinity states of channel, respectively. The sampling rate for the simulation of the binding transitions was 10 Hz. (D) The percentage of high-affinity channels (CH and CHTx) in the whole system calculated by kinetic model. (E) The percentage of the induced-fit pathway over the total (induced fit and conformational selection) pathway for binding. The preference was calculated by using the kinetic model.
In the simulated binding transitions (lower plot in Fig. 4C), we can discriminate the high-affinity from the low-affinity states and elucidate that the fraction of the high-affinity states increased with an increasing AgTx2 concentration (Fig. 4D). The fractions of the high-affinity channel (Fig. 4D) were quite similar to those of the experimental and simulated Pbound (Fig. 4B), indicating that most of the AgTx2-bound state was a high-affinity state (CHTx). The AgTx2-bound low-affinity state (CLTx) and AgTx2-unbound high-affinity state (CHTx) appeared intermittently (Fig. 4C, bottom plot).
In our four-state model (Fig. 4A), if AgTx2 was bound to the channel, then the rate of change from the low- to high-affinity channels (CLTx to CHTx) occurred 400 times faster than without AgTx2 binding (CL to CH), indicating that the binding of AgTx2 induced a change in channel affinity from low to high during binding. This change in affinity was probably related to the change in channel structure, as discussed later.
Induced fit is the dominant pathway of AgTx2 binding
Generally, there are two pathways of ligand-receptor binding that account for affinity changes: conformational selection and induced fit (17–21, 23, 44). For conformational selection, the receptor presents different structures with different affinities to the ligand without binding of the ligand, and the ligand preferentially binds to the high-affinity state of the receptor (CL ↔ CH ↔ CHTx). By contrast, in the induced-fit pathway, the receptor adopts a low-affinity state before the ligand binds and changes its structure to the high-affinity state during ligand binding (CL ↔ CLTx ↔ CHTx). It is generally difficult to distinguish these two pathways by macroscopic analyses. Here, we obtained a relevant four-state model (Fig. 4A) deduced from single-molecule AFM measurements, which enabled us to distinguish the dominant binding pathway.
To determine the dominant binding pathway of AgTx2 at the K+ channel, we calculated the fraction of the induced-fit pathway over the total (conformational selection plus induced-fit pathways) using the rate constants via the following equation: k34P3/(k34P3 + k12P1) (see Materials and Methods for their definitions) (45). Although the fraction was slightly affected by the AgTx2 concentration, almost all the binding events occurred via the induced-fit pathway (Fig. 4E), indicating that the dynamics of binding between the K+ channel and AgTx2 could be described by this mechanism. We also confirmed this by calculating the AgTx2 concentration dependency of kobs (fig. S4). Our single-molecule study revealed that the induced-fit pathway almost exclusively describes AgTx2-KcsA binding.
Noninactivating channels are always in a state of high affinity for AgTx2
According to previous structural investigations of the binding of AgTx2 to the KcsA channel (11, 12, 16), the side chain of AgTx2 residue K27, which is highly conserved across the scorpion toxin families, interacts with the selectivity filter (around sites known as S4 and S5) of the channel. It is known that the structure in the selectivity filter region adopts at least two conformational states: the straight (conductive) form (14) and the bending (collapsed, inactivated) (46) form of the filter (Fig. 5A). Lange et al. (47, 48) have reported that the scorpion toxin kaliotoxin (KTX), which also has K27, induces the conformational change in the selectivity filter of the KcsA-Kv1.3 chimera channel from the inactivated to the conductive form. This phenomenon suggests that the induced-fit dynamics observed here were closely related to the structural change around the selectivity filter.
Fig. 5. Binding dynamics of the noninactivating channel.
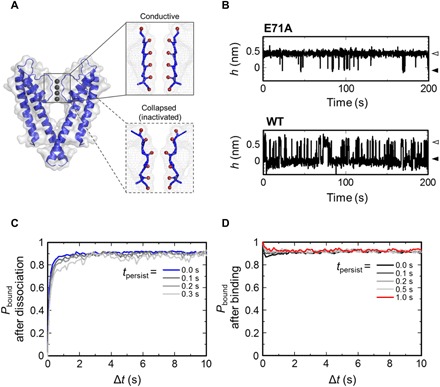
(A) Side view of the KcsA channel and enlarged view of the selectivity filter region with the conductive state (PDB ID: 1K4C) and the collapsed state (1K4D). (B) Height transitions of the E71A and WT channels upon AgTx2 binding. White and black arrowheads indicate the AgTx2-bound and AgTx2-unbound states of the channels, respectively. The noninactivating mutant (E71A) showed markedly high affinity to AgTx2 compared with WT. The binding probabilities of AgTx2 to the E71A mutant obtained by HS-AFM imaging and simulation with only the high-affinity channel are plotted in Fig. 4B. (C and D) Time course of Pbound immediately after or for a persistence of up to 1 or 0.3 s from the dissociation (C) and the binding (D) events. Δt is the time elapsed from the end of tpersist. The height transitions used in this analysis were measured in 10 mM Hepes (pH 7.5) containing 200 mM KCl and 20 nM AgTx2.
To investigate the effect of the structure around the selectivity filter on AgTx2 binding, we also observed the E71A mutant. (The KcsA channel with Q58A/T61S/R64D mutations, which has been described thus far, is referred to as “WT” hereafter for simplicity.) Because the E71A mutation stabilizes the conductive form (49, 50), the E71A mutant is known as a noninactivating mutant. In addition, the side chain of D80 usually orients inside the channel, but in the case of the E71A mutant, the carboxylate side chain of D80 is flipped out to the extracellular surface of the channel (49, 50). Figure 5B shows the height transitions of the E71A and WT channels upon AgTx2 binding. The E71A mutant readily bound to AgTx2—the experimental Pbound value was 0.94 for the E71A mutant, which is six times greater than that of the WT (Fig. 4B). The theoretical Pbound value, calculated with only the AgTx2-bound and AgTx2-unbound states of the high-affinity channel (CH and CHTx), was 0.97 at 100 nM AgTx2, which was almost the same as the experimental Pbound of E71A. These results support the idea that the conductive form of the selectivity filter corresponds to the high-affinity state.
If the noninactivating channel always adopts the high-affinity state, the induced-fit dynamics will be diminished. We examined whether the affinity change after binding and dissociation events diminish using event-oriented analysis (Fig. 5, C and D). The persistence time from binding and dissociation events had almost no effect on Pbound(Δt), indicating that the binding dynamics of the E71A mutant were mostly described by a two-state (AgTx2-bound and AgTx2-unbound) model; thus, the affinity change was not required to explain the dynamics. These results suggest that the noninactivating channel is always in a high-affinity state for AgTx2, inferring that the conductive state of the selectivity filter corresponds to the high-affinity state of the channel.
DISCUSSION
We successfully observed the dynamics of binding of AgTx2 to a K+ channel by HS-AFM. Single-molecule analysis indicated that the AgTx2 increases the affinity of the K+ channel to AgTx2 as binding persists, and conversely, the affinity decreases after AgTx2 leaves the channel. A four-state model, including high- and low-affinity states of the channel, explained the experimental data. The induced-fit pathway is the almost-exclusive mode for the binding dynamics of AgTx2. On the other hand, the noninactivating channel always showed high affinity for AgTx2, implying that the AgTx2-induced affinity change corresponded to structural changes around the selectivity filter.
HS-AFM observations revealed the induced-fit dynamics upon AgTx2 binding (Fig. 3), which are tightly coupled to the structural state of the selectivity filter of the channel (Fig. 5). This induced fit might be similar to KTX (47), and the conductive state of the selectivity filter corresponds to the high-affinity state. One possible explanation for our observations is that D80 of the channel flips out to face R24 of the AgTx2 during induced fit, and the flipped structure corresponds to the high-affinity state. If D80 is flipped, the distance between D80 of the channel and R24 of AgTx2 becomes shorter (~0.5 nm), and the two residues can readily bind. Using the event-oriented analysis, a calculation of Boltzmann’s H function, showing the change of entropy over time, showed that the binding process is mainly driven by energetic forces (fig. S5), confirming this idea.
Another possible explanation for the structural change upon induced fit is that the channel loses fourfold symmetry upon binding of the asymmetric AgTx2 molecule. Bhate and McDermott (50) reported that the E71A mutation increases the structural disorder of the selectivity filter. This finding indicates that the structures around the selectivity filter of the E71A mutant readily lose the fourfold symmetry. The induced fit from the low- to high-affinity states takes approximately 260 ms (Fig. 4A), indicating that the structural change is not very fast. This change might occur not only within a small space (around only the selectivity filter) but also within a large space (loosening the fourfold symmetry of the channel) during AgTx2 binding. If a very long molecular dynamics (MD) simulation can be carried out in the future, we could confirm whether these dynamics occur upon AgTx2 binding.
We also observed dynamic disorder in the system (51, 52) (movie S2 and fig. S6). The Pbound of some channels changed over time; for example, Pbound of channel a in fig. S6 changed markedly at 60 and 100 s, and that of channel f changed at 100 s. These transitions were not observed frequently, implying that the time constants of such transitions appear to be longer than the observation time and are difficult to estimate. These transitions remind us of the mode switch of the KcsA channel, in which channel gating spontaneously turns to different modes with high and low open probability (53), while transition rates between the modes remain elusive. Accordingly, the individual Pbound values for each channel have a distribution (fig. S6). This phenomenon indicates that rate constants differ among channels and also change in time even for each channel. In this study, there was no obvious correlation between the interchannel distance and individual Pbound value (fig. S6). It would be very interesting in further analyses to clarify the structural states that affect the fluctuation of reaction rates and individual Pbound values. Note that the rate constants that we obtained correspond to the time-averaged values.
We revealed that the induced-fit pathway is dominant in the AgTx2-KcsA system. Physiological implications of the induced-fit mechanism include potentially enhanced fidelity of molecular recognition. Moreover, the channel affinity remained high for a while after unbinding. This “memory” may create spatial inhomogeneity of channels. Such sophisticated spatial correlation of toxin binding to channels is a future issue to be examined with HS-AFM.
Our strategy to evaluate the rate constants was to use the event-oriented analysis that we developed recently (42, 43). This analysis is easily applied to the time-series data obtained by single-molecule observations. In addition, this method can be used to estimate rate constants with approximately 10 times fewer data than the dwell time analysis and with similar or somewhat more statistics compared to the correlation function (fig. S7). Moreover, the event-oriented analysis has two advantages over analysis based on the correlation function: (i) this analysis is very intuitive and should be familiar to researchers, since time-dependent quantities from a certain stimulus (event), such as temperature jump or light emission, are usually measured in experiments and (ii) it is easy to introduce a persistent time from the event (tpersist) or to compare the results to other events (42), which enables us to investigate the effect of such an event on the dynamics. The event-oriented analysis is applicable for time-series data obtained by any measurement method, but the current time resolution of the event-oriented analysis using HS-AFM data is limited by the sampling rate of HS-AFM (10 fps). Further improvement of HS-AFM instruments to achieve better time resolution would expand the application of event-oriented analysis to analyzing faster biological dynamics.
In this paper, we demonstrate that single-molecule HS-AFM can be used not only as a visualization tool for single-molecule events but also as an advanced analytic tool. Recently, HS-AFM machines have become more popular, and the stochastic analysis (event-oriented analysis) carried out in this study can thus be widely applicable to various types of biological molecules. We believe that our study can facilitate studies of the binding dynamics of a wide variety of biological molecules, including antigen-antibody, ligand-receptor, and DNA-modifier protein interactions by HS-AFM.
MATERIALS AND METHODS
Materials
The KcsA channel was expressed in Escherichia coli BL21(DE3)pLysS cells, solubilized with n-dodecyl β-d-maltopyranoside (DDM; Dojindo), and purified as previously described (54). We genetically deleted the cytoplasmic domain (126 to 160) to reduce lateral fluctuation of the channel on the AFM substrate. To mimic the toxin-binding surface of the Shaker K+ channel, we introduced three mutations (Q58A, T61S, and R64D). All the channels used in this study carried this Shaker K+ channel–mimicking mutation, and thus, we refer to this triple mutant as “WT” herein for simplicity. To immobilize the channel onto a Ni2+-coated mica surface (41, 55), a hexahistidine tag was introduced at the C terminus (as residues 126 to 131). AgTx2 and DMPC were purchased from Smartox Biotechnology (Saint-Egrève, France) and Avanti Polar Lipids (Alabaster, Alabama), respectively.
HS-AFM observations
The KcsA channels were reconstituted into DMPC bilayers on a mica substrate with their extracellular surface facing outward, using a previously developed method (41). To suppress lateral diffusion of the channels, we attached the channels to the mica surface through a Ni2+–His-tag interaction. First, the mica surface was covered with 0.5 M NiCl2 solution for 5 min and rinsed with pure water to coat the surface with Ni2+ (55). Second, the addition of solubilized channel (10 μg/ml in 10 mM Hepes, 2 M KCl, and 0.06% DDM) onto the Ni2+-coated surface for 5 min provided a channel-decorated surface. Last, DMPC liposome solution (50 μg/ml) containing 120 μM DDM was applied to the channel-decorated surface to reconstitute the channel into the bilayer with the toxin-binding site exposed. The imaging buffer was 10 mM Hepes (pH 7.5) and 100 or 200 mM KCl (see figure legends). We used AFM data with 100 mM KCl only in Fig. 2 because a low KCl concentration results in a slower dissociation rate. A typical example of the binding transition was observed. AgTx2 was added to the imaging buffer in the concentration range from 10 to 1000 nM. All experiments were performed at 25°C under equilibrium conditions, that is, more than 10 min after AgTx2 application. We confirmed that the system was at equilibrium using surface plasmon resonance measurements. For HS-AFM observations, a laboratory-built HS-AFM was used (30, 31). The cantilever used was AC7 (Olympus Co., Tokyo, Japan) with an electron beam deposition tip. The typical resonant frequency and quality factor in water of AC7 are 700 kHz and 2, respectively.
Extraction of the height transition from HS-AFM data
We extracted height transitions using a self-written program based on Igor Pro (Wavemetrics Inc.). First, lateral displacement of the channels due to thermal drift was compensated for using an image correlation algorithm, and then the average height around the center of the extracellular surface (approximately 20 pixels, corresponding to approximately 0.6 nm2) of the channel was measured. We used height transitions from 86 channels for the analysis. Baseline fluctuations in the raw data were removed (fig. S8) according to the calculations detailed in the Supplementary Materials.
Event-oriented analysis
Time series of AFM measurements, h(t), were assigned to two hypothesized states—AgTx2-bound and AgTx2-unbound—according to the AFM surface height threshold. Hence, two transitions (events) were observed: from bound to unbound and from unbound to bound. Instead of the previous analyses using autocorrelation function and dwell time (56, 57), the event-oriented analysis for determining the time course of Pbound(Δt) was applied to the HS-AFM measurements and the bound-unbound transitions were observed at 20 nM. First, fbound(h; Δt) was defined as the distribution of the height from the moment of AgTx2 binding, where Δt is the time from the moment of binding; that is, Δt is zero when AgTx2 binds. All binding events were collected from the measurements to evaluate fbound(h; Δt); that is, fbound(h; Δt) represents an ensemble average of trajectories of height. The number of binding events in our dataset was 6514. Then, Pbound was estimated using the following equation
Similarly, another time course of the binding probability Punbound(Δt) was defined, in which Δt is the time from the moment of dissociation. We used this description for subsequent equations in this section and described Punbound(Δt) as “Pbound(Δt) from dissociation” in the remaining text and figures. The number of dissociation events was 6511. Neither Pbound(Δt) nor Punbound(Δt) were single-exponential events, as described in the text earlier, indicating the presence of more than two states in the system. To clarify this phenomenon, a persistence time in a state was introduced, tpersist, which is the time spent in the bound or unbound state without transition. Here, Pbound(Δt; tpersist) was defined as follows
Here, Δt is the time after the persistent residence in a state. Thus, Pbound(Δt; tpersist) is closely related to a three-time correlation function, which is known to be a powerful tool to reveal the detailed dynamics of complicated systems, such as biomolecules (57–60). Punbound(Δt; tpersist) was defined in the same manner.
Estimation of rate constants
According to the event-oriented analysis, there are two conformational states in the channel: high- and low-affinity states, denoted as CH and CL, respectively. Considering the binding of AgTx2, four states were expected to exist: CH (state 1; this numbering was used in the mathematical treatment below), CL (state 2), CHTx (state 3), and CLTx (state 4) (Fig. 4A). The four-state model was also supported by the conventional dwell time analysis, in which both bound and unbound dwell time distribution could be fitted by double-exponential functions (fig. S2, A and B). (A three-state kinetic model was also tested but did not successfully reproduce the measurements: In particular, the model could not reproduce a sudden decrease of Pbound when tpersist = 0 s in Fig. 3E since the model lacks CLTx.) The rate from state i to j is denoted by kij. The time courses of the probabilities of states were calculated by solving the following simultaneous differential equation
where Pi is the probability of state i and . Two transitions describing AgTx2 binding (state 1 → 3 and 2 → 4) were considered to be proportional to the concentration of the toxin ([AgTx2]). Note that among eight rate constants, one is a dependent variable because of the detailed balance, for example, k43 = k34k42k21k13/k31k12k24.
To estimate the rate constants, four time courses of binding probability were used to fit simultaneously: Pbound(Δt; tpersist = 0 s), Pbound(Δt; tpersist = 1.0 s), Punbound(Δt; tpersist = 0 s), and Punbound(Δt; tpersist = 1.5 s). Pbound(Δt; tpersist) did not greatly alter tpersist > 1.0 s, and Punbound(Δt; tpersist) did not greatly change tpersist > 1.5 s. P3 plus P4 [P3 + 4(Δt)] is the binding probability, which is measured by AFM.
The rates were optimized using the least-squares method, in which the following function (E) was minimized
where weights w of 10 for Δt < 0.5 s, 2 for 0.5 s ≤ Δt < 5 s, and 1 otherwise were used to precisely fit the sudden changes on the short time scale. In addition, two scaling parameters (s1 = 0.9 and s2 = 0.75) were used to determine the best fit. These four curves were fitted by solving the above simultaneous differential equations with different initial conditions of the probability: Pbound(Δt; tpersist = 0 s) was calculated by setting P4(0) = 0.65 and P3(0) = 0.35. Pbound(Δt; tpersist = 1.0 s) was calculated by setting P3(0) = 1, assuming all channels adopt a high-affinity state if a long time passes after the binding of AgTx2. Similarly, Punbound(Δt; tpersist = 1.5 s) was calculated by setting P2(0) = 1, assuming that all channels adopt a low-affinity state if a long time passes after the dissociation of AgTx2. Punbound(Δt; tpersist = 0 s) was calculated by setting P1(0) = 0.35 and P2(0) = 0.65. All the rates were limited to less than 100 s−1 in the fitting because AFM cannot measure binding events faster than 100 s−1; AFM requires at least 10 ms to scan the entire surface of the AgTx2 molecule. The resultant fitted curves, in addition to the measured ones, are shown in fig. S2 (C and D). The rates were evaluated at 20 nM AgTx2 because the largest amount of data was obtained at that concentration.
Supplementary Material
Acknowledgments
We thank M. Iwamoto (University of Fukui), F. Osawa (Keio University), N. Kodera (Kanazawa University), M. Shibata (Kanazawa University), and T. Ando (Kanazawa University) for fruitful discussions and for sharing laboratory instruments. A.S. thanks S. Morita (Kanazawa University) and C. Miller (Brandeis University) for reading and providing comments on this paper. A.S. also thanks M. Yamatake for experimental assistance. Funding: A.S. thanks PRESTO (JST), Grant-in-Aid for Young Scientists (A) (17H05058), and Challenging Research (Exploratory) (18K19422) for funding. T.S. thanks Grant-in-Aid for Young Scientists (B) (17K17768) for funding. S.O. thanks Grant-in-Aid for Scientific Research (16H00759 and 17H04017). The calculations for the event-oriented analysis and fitting of rate constants were carried out on the supercomputers at the Research Center for Computational Science in Okazaki, Japan. Author contributions: A.S. and S.O. designed the study. A.S. performed the experiments. T.U. provided training in the HS-AFM observation technique and basic image analysis for A.S and wrote the image tracking and height extraction procedures. T.S. wrote the baseline correction program for height transitions, performed the event-oriented analysis, and calculated the rate constants. All authors wrote the paper. Competing interests: The authors declare that they have no competing interests. Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Additional data related to this paper may be requested from the authors.
SUPPLEMENTARY MATERIALS
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/5/7/eaax0495/DC1
Fig. S1. Binding transitions obtained by HS-AFM.
Fig. S2. Dwell time distributions and the fitted time course of Pbound.
Fig. S3. Simulated binding transitions.
Fig. S4. The AgTx2 concentration dependence on kobs.
Fig. S5. Boltzmann’s H function.
Fig. S6. Dynamic disorder of binding transitions and correlation of individual Pbound values and interchannel distance.
Fig. S7. Dwell time distributions obtained by dwell time analysis, event-oriented analysis, and correlation function.
Fig. S8. Removing fluctuations in the baseline.
Movie S1. HS-AFM movie of AgTx2 binding on the KcsA channels.
Movie S2. HS-AFM movie showing dynamic disorder of binding transitions.
Reference (61)
REFERENCES AND NOTES
- 1.B. Hille, Ion channels of excitable membranes (Sinauer Associates Inc, Sunderland, ed. 3, 2001). [Google Scholar]
- 2.Iwamoto M., Oiki S., Constitutive boost of a K+ channel via inherent bilayer tension and a unique tension-dependent modality. Proc. Natl. Acad. Sci. U.S.A. 115, 13117–13122 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.F. M. Ashcroft, Ion Channels and Disease: Channelopathies (Academic Press, 2000). [Google Scholar]
- 4.Possani L. D., Merino E., Corona M., Bolivar F., Becerril B., Peptides and genes coding for scorpion toxins that affect ion-channels. Biochimie 82, 861–868 (2000). [DOI] [PubMed] [Google Scholar]
- 5.Possani L. D., Becerril B., Delepierre M., Tytgat J., Scorpion toxins specific for Na+-channels. Eur. J. Biochem. 264, 287–300 (1999). [DOI] [PubMed] [Google Scholar]
- 6.Tytgat J., Chandy K. G., Garcia M. L., Gutman G. A., Martin-Eauclaire M.-F., van der Walt J. J., Possani L. D., A unified nomenclature for short-chain peptides isolated from scorpion venoms: α-KTx molecular subfamilies. Trends Pharmacol. Sci. 20, 444–447 (1999). [DOI] [PubMed] [Google Scholar]
- 7.Carbone E., Wanke E., Prestipino G., Possani L. D., Maelicke A., Selective blockage of voltage-dependent K+ channels by a novel scorpion toxin. Nature 296, 90–91 (1982). [DOI] [PubMed] [Google Scholar]
- 8.Miller C., The charybdotoxin family of K+ channel-blocking peptides. Neuron 15, 5–10 (1995). [DOI] [PubMed] [Google Scholar]
- 9.Kuzmenkov A. I., Grishin E. V., Vassilevski A. A., Diversity of potassium channel ligands: Focus on scorpion toxins. Biochemistry 80, 1764–1799 (2015). [DOI] [PubMed] [Google Scholar]
- 10.Garcia M. L., Garcia-calvo M., Hidalgo L. P., Lee A., Mackinnon R., Purification and characterization of three inhibitors of voltage-dependent K+ channels from leiurus quinquestriatus var. hebraeus venom. Biochemistry 33, 6834–6839 (1994). [DOI] [PubMed] [Google Scholar]
- 11.MacKinnon R., Cohen S. L., Kuo A., Lee A., Chait B. T., Structural conservation in prokaryotic and eukaryotic potassium channels. Science 280, 106–109 (1998). [DOI] [PubMed] [Google Scholar]
- 12.Eriksson M. A. L., Roux B., Modeling the structure of agitoxin in complex with the Shaker K+ channel: A computational approach based on experimental distance restraints extracted from thermodynamic mutant cycles. Biophys. J. 83, 2595–2609 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schrempf H., Schmidt O., Kümmerlen R., Hinnah S., Müller D., Betzler M., Steinkamp T., Wagner R., A prokaryotic potassium ion channel with two predicted transmembrane segments from Streptomyces lividans. EMBO J. 14, 5170–5178 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Doyle D. A., Cabral J. M., Pfuetzner R. A., Kuo A., Gulbis J. M., Cohen S. L., Chait B. T., MacKinnon R., The structure of the potassium channel: Molecular basis of K+ conduction and selectivity. Science 280, 69–77 (1998). [DOI] [PubMed] [Google Scholar]
- 15.Krezel A. M., Kasibhatla C., Hidalgo P., Mackinnon R., Wagner G., Solution structure of the potassium channel inhibitor agitoxin 2: Caliper for probing channel geometry. Protein Sci. 4, 1478–1489 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Takeuchi K., Yokogawa M., Matsuda T., Sugai M., Kawano S., Kohno T., Nakamura H., Takahashi H., Shimada I., Structural basis of the KcsA K+ channel and agitoxin2 pore-blocking toxin interaction by using the transferred cross-saturation method. Structure 11, 1381–1392 (2003). [DOI] [PubMed] [Google Scholar]
- 17.Vogt A. D., Di Cera E., Conformational selection or induced fit? A critical appraisal of the kinetic mechanism. Biochemistry 51, 5894–5902 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Paul F., Weikl T. R., How to distinguish conformational selection and induced fit based on chemical relaxation rates. PLOS Comput. Biol. 12, e1005067 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gianni S., Dogan J., Jemth P., Distinguishing induced fit from conformational selection. Biophys. Chem. 189, 33–39 (2014). [DOI] [PubMed] [Google Scholar]
- 20.Changeux J.-P., Edelstein S., Conformational selection or induced-fit? 50 years of debate resolved. F1000 Biol. Rep. 3, 19 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Csermely P., Palotai R., Nussinov R., Induced fit, conformational selection and independent dynamic segments: An extended view of binding events. Trends Biochem. Sci. 35, 539–546 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kovermann M., Grundström C., Sauer-Eriksson A. E., Sauer U. H., Wolf-Watz M., Structural basis for ligand binding to an enzyme by a conformational selection pathway. Proc. Natl. Acad. Sci. U.S.A. 114, 6298–6303 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Silva D.-A., Bowman G. R., Sosa-Peinado A., Huang X., A role for both conformational selection and induced fit in ligand binding by the LAO protein. PLOS Comput. Biol. 7, e1002054 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Peuker S., Cukkemane A., Held M., Noé F., Kaupp U. B., Seifert R., Kinetics of ligand-receptor interaction reveals an induced-fit mode of binding in a cyclic nucleotide-activated protein. Biophys. J. 104, 63–74 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.C. Miller, Ion Channel Reconstitution (Springer Science+Business Media, 1986). [Google Scholar]
- 26.Kim J.-Y., Kim C., Lee N. K., Real-time submillisecond single-molecule FRET dynamics of freely diffusing molecules with liposome tethering. Nat. Commun. 6, 6992 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gadkari V. V., Harvey S. R., Raper A. T., Chu W.-T., Wang J., Wysocki V. H., Suo Z., Investigation of sliding DNA clamp dynamics by single-molecule fluorescence, mass spectrometry and structure-based modeling. Nucleic Acids Res. 46, 3103–3118 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Park J., Jeon Y., in D., Fishel R., Ban C., Lee J.-B., Single-molecule analysis reveals the kinetics and physiological relevance of MutL-ssDNA binding. PLOS ONE. 5, e15496 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Luo Y., North J. A., Poirier M. G., Single molecule fluorescence methodologies for investigating transcription factor binding kinetics to nucleosomes and DNA. Methods 70, 108–118 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ando T., Kodera N., Takai E., Maruyama D., Saito K., Toda A., A high-speed atomic force microscope for studying biological macromolecules. Proc. Natl. Acad. Sci. U.S.A. 98, 12468–12472 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Uchihashi T., Kodera N., Ando T., Guide to video recording of structure dynamics and dynamic processes of proteins by high-speed atomic force microscopy. Nat. Protoc. 7, 1193–1206 (2012). [DOI] [PubMed] [Google Scholar]
- 32.Ando T., Uchihashi T., Scheuring S., Filming biomolecular processes by high-speed atomic force microscopy. Chem. Rev. 114, 3120–3188 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kodera N., Yamamoto D., Ishikawa R., Ando T., Video imaging of walking myosin V by high-speed atomic force microscopy. Nature 468, 72–76 (2010). [DOI] [PubMed] [Google Scholar]
- 34.Uchihashi T., Iino R., Ando T., Noji H., High-speed atomic force microscopy reveals rotary catalysis of rotorless F1-ATPase. Science 333, 755–758 (2011). [DOI] [PubMed] [Google Scholar]
- 35.Shibata M., Nishimasu H., Kodera N., Hirano S., Ando T., Uchihashi T., Nureki O., Real-space and real-time dynamics of CRISPR-Cas9 visualized by high-speed atomic force microscopy. Nat. Commun. 8, 1430 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ruan Y., Miyagi A., Wang X., Chami M., Boudker O., Scheuring S., Direct visualization of glutamate transporter elevator mechanism by high-speed AFM. Proc. Natl. Acad. Sci. 114, 1584–1588 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shibata M., Yamashita H., Uchihashi T., Kandori H., Ando T., High-speed atomic force microscopy shows dynamic molecular processes in photoactivated bacteriorhodopsin. Nat. Nanotechnol. 5, 208–212 (2010). [DOI] [PubMed] [Google Scholar]
- 38.Ruan Y., Kao K., Lefebvre S., Marchesi A., Corringer P.-J., Hite R. K., Scheuring S., Structural titration of receptor ion channel GLIC gating by HS-AFM. Proc. Natl. Acad. Sci. U.S.A. 115, 10333–10338 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mori T., Sugiyama S., Byrne M., Johnson C. H., Uchihashi T., Ando T., Revealing circadian mechanisms of integration and resilience by visualizing clock proteins working in real time. Nat. Commun. 9, 3245 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yamamoto D., Ando T., Chaperonin GroEL–GroES functions as both alternating and non-alternating engines. J. Mol. Biol. 428, 3090–3101 (2016). [DOI] [PubMed] [Google Scholar]
- 41.Sumino A., Uchihashi T., Oiki S., Oriented reconstitution of the full-length KcsA potassium channel in a lipid bilayer for AFM imaging. J. Phys. Chem. Lett. 8, 785–793 (2017). [DOI] [PubMed] [Google Scholar]
- 42.Sumikama T., Oiki S., Digitalized K+ occupancy in the nanocavity holds and releases queues of K+ in a channel. J. Am. Chem. Soc. 138, 10284–10292 (2016). [DOI] [PubMed] [Google Scholar]
- 43.Sumikama T., Saito S., Ohmine I., Mechanism of ion permeation through a model channel: Roles of energetic and entropic contributions. J. Chem. Phys. 139, 165106 (2013). [DOI] [PubMed] [Google Scholar]
- 44.Hatzakis N. S., Single molecule insights on conformational selection and induced fit mechanism. Biophys. Chem. 186, 46–54 (2014). [DOI] [PubMed] [Google Scholar]
- 45.Hammes G. G., Chang Y.-C., Oas T. G., Conformational selection or induced fit: A flux description of reaction mechanism. Proc. Natl. Acad. Sci. U.S.A. 106, 13737–13741 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhou Y., Morais-Cabral J. H., Kaufman A., MacKinnon R., Chemistry of ion coordination and hydration revealed by a K+ channel-Fab complex at 2.0 Å resolution. Nature 414, 43–48 (2001). [DOI] [PubMed] [Google Scholar]
- 47.Lange A., Giller K., Hornig S., Martin-Eauclaire M.-F., Pongs O., Becker S., Baldus M., Toxin-induced conformational changes in a potassium channel revealed by solid-state NMR. Nature 440, 959–962 (2006). [DOI] [PubMed] [Google Scholar]
- 48.Zachariae U., Schneider R., Velisetty P., Lange A., Seeliger D., Wacker S. J., Karimi-Nejad Y., Vriend G., Becker S., Pongs O., Baldus M., de Groot B. L., The molecular mechanism of toxin-induced conformational changes in a potassium channel: Relation to C-type inactivation. Structure 16, 747–754 (2008). [DOI] [PubMed] [Google Scholar]
- 49.Cordero-Morales J. F., Cuello L. G., Zhao Y., Jogini V., Cortes D. M., Roux B., Perozo E., Molecular determinants of gating at the potassium-channel selectivity filter. Nat. Struct. Mol. Biol. 13, 311–318 (2006). [DOI] [PubMed] [Google Scholar]
- 50.Bhate M. P., McDermott A. E., Protonation state of E71 in KcsA and its role for channel collapse and inactivation. Proc. Natl. Acad. Sci. U.S.A. 109, 15265–15270 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Karplus M., Aspects of protein reaction dynamics: Deviations from simple behavior. J. Phys. Chem. B 104, 11–27 (2000). [Google Scholar]
- 52.English B. P., Min W., van Oijen A. M., Lee K. T., Luo G., Sun H., Cherayil B. J., Kou S. C., Xie X. S., Ever-fluctuating single enzyme molecules: Michaelis-Menten equation revisited. Nat. Chem. Biol. 2, 87–94 (2006). [DOI] [PubMed] [Google Scholar]
- 53.Chakrapani S., Cordero-Morales J. F., Perozo E., A quantitative description of KcsA gating II: Single-channel currents. J. Gen. Physiol. 130, 479–496 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Iwamoto M., Shimizu H., Inoue F., Konno T., Sasaki Y. C., Oiki S., Surface structure and its dynamic rearrangements of the KcsA potassium channel upon gating and tetrabutylammonium blocking. J. Biol. Chem. 281, 28379–28386 (2006). [DOI] [PubMed] [Google Scholar]
- 55.Hansma H. G., Laney D. E., Bezanilla M., Sinsheimer R. L., Hansma P. K., Applications for atomic force microscopy of DNA. Biophys. J. 68, 1672–1677 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ball F. G., Kerry C. J., Ramsey R. L., Sansom M. S., Usherwood P. N., The use of dwell time cross-correlation functions to study single-ion channel gating kinetics. Biophys. J. 54, 309–320 (1988). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rothberg B. S., Bello R. A., Magleby K. L., Two-dimensional components and hidden dependencies provide insight into ion channel gating mechanisms. Biophys. J. 72, 2524–2544 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ono J., Takada S., Saito S., Couplings between hierarchical conformational dynamics from multi-time correlation functions and two-dimensional lifetime spectra: Application to adenylate kinase. J. Chem. Phys. 142, 212404 (2015). [DOI] [PubMed] [Google Scholar]
- 59.Mori T., Saito S., Dynamic heterogeneity in the folding/unfolding transitions of FiP35. J. Chem. Phys. 142, 135101 (2015). [DOI] [PubMed] [Google Scholar]
- 60.Mori T., Saito S., Molecular mechanism behind the fast folding/unfolding transitions of villin headpiece subdomain: Hierarchy and heterogeneity. J. Phys. Chem. B 120, 11683–11691 (2016). [DOI] [PubMed] [Google Scholar]
- 61.R. Kubo, M. Toda, N. Hashitsume, Statistical Physics II : Nonequilibrium Statistical Mechanics (Springer Berlin Heidelberg, 1991). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/5/7/eaax0495/DC1
Fig. S1. Binding transitions obtained by HS-AFM.
Fig. S2. Dwell time distributions and the fitted time course of Pbound.
Fig. S3. Simulated binding transitions.
Fig. S4. The AgTx2 concentration dependence on kobs.
Fig. S5. Boltzmann’s H function.
Fig. S6. Dynamic disorder of binding transitions and correlation of individual Pbound values and interchannel distance.
Fig. S7. Dwell time distributions obtained by dwell time analysis, event-oriented analysis, and correlation function.
Fig. S8. Removing fluctuations in the baseline.
Movie S1. HS-AFM movie of AgTx2 binding on the KcsA channels.
Movie S2. HS-AFM movie showing dynamic disorder of binding transitions.
Reference (61)


