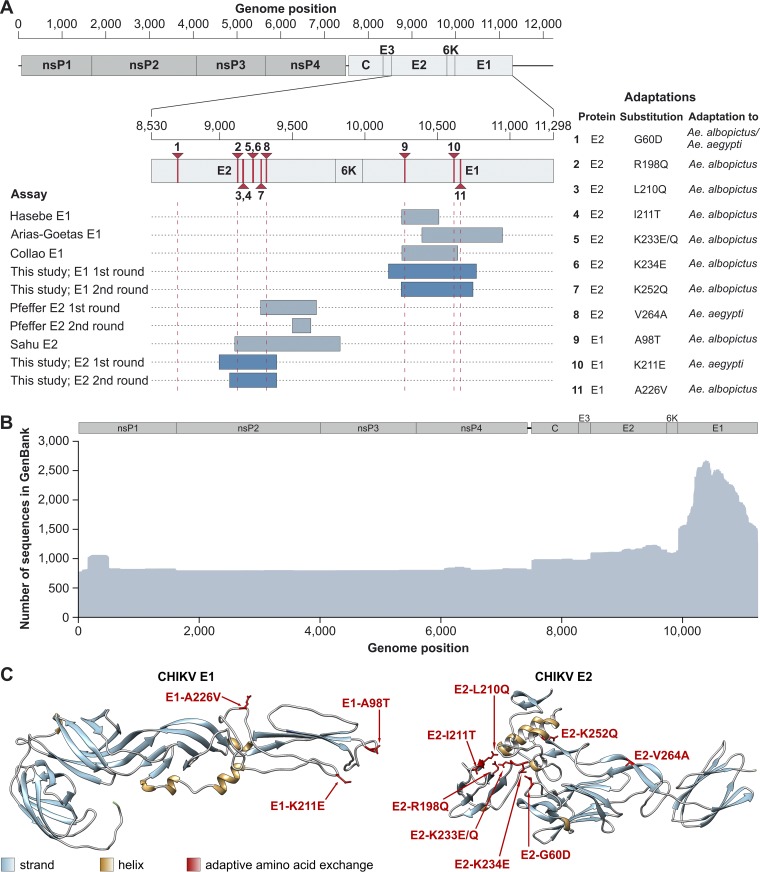
FIG 1.
Adaptive amino acid exchanges in CHIKV and assay design. (A) Genomic localization of adaptive mutations and commonly used PCR assays. Genome positions are shown for a CHIKV reference genome (GenBank accession no. MG967666). Positions of adaptive mutations are indicated in red and by vertical dotted red lines at their extremities. (B) Sequence coverage and identity of the coding region of the CHIKV genome based on all available GenBank entries (18 March 2019). Coverage and identity were calculated using Geneious 9.1.8 and were visualized using R version 3.5.2. (C) Tridimensional localization of known adaptive amino acid exchanges in the CHIKV E1 and E2 proteins. The amino acid exchanges E1-T98A (7), E1-K211E (13), E1-A226V (7), E2-G60D (9), E2-R198Q (7), E2-L210Q (7), E2-I211T (9), E2-K233E/Q (7), E2-K234E (7), E2-K252Q (7), and E2-V264A (12) are depicted.