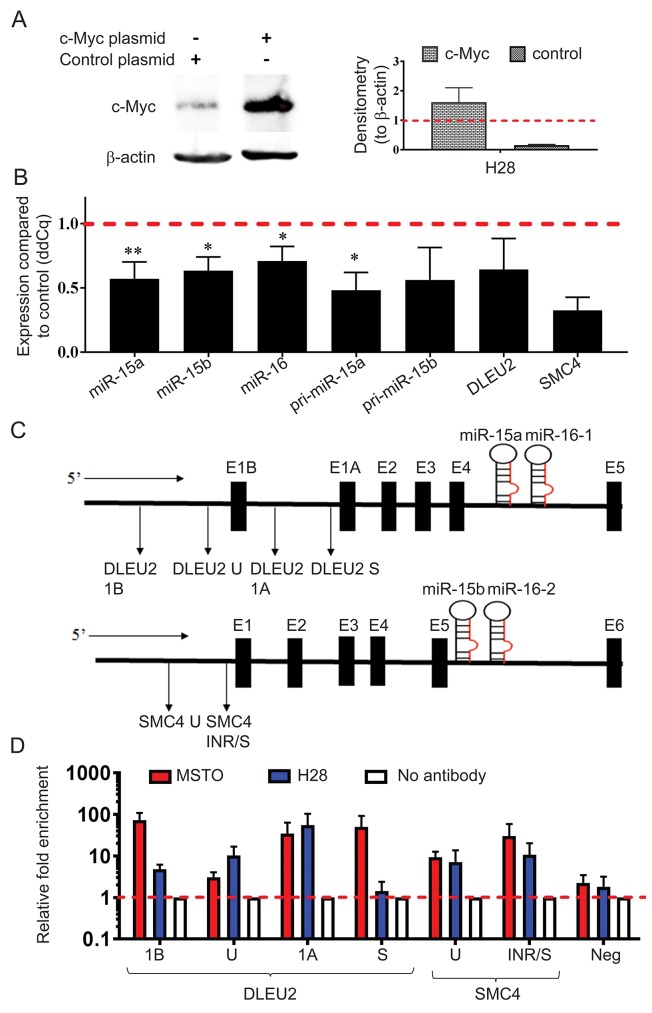
Figure 5. MYC overexpression causes downregulation of miR-15b/16-2 and miR-15a/16-1 and their host genes via binding to the DLEU2 and SMC4 promoters.
(A) H28 cells were transfected with pcDNA3.1(+) and pcDNA3.1(+)MYC plasmids and upregulation of Myc protein expression was confirmed via western blotting with β-actin included as a loading control and normalizer for densitometry. Data are the mean of 3 independent experiments ±SD. (B) Mature miRNA, pri-miRNA and host gene expression was analyzed using RT-qPCR with RNU6B and 18S as reference genes. MYC construct transfection induced downregulation of mature, pri- and host gene mRNAs from both the miR-15a/16-1 and miR-15b/16-2 clusters compared to vector control-transfected cells. Data are the mean of 3 independent experiments ±SD. * = p ≤ 0.05; ** = p ≤ 0.01. (C) Schematic representation of the promoter regions of the host genes DLEU2 and SMC4 with the amplicon locations used for ChIP analysis indicated below each gene. (D) ChIP was performed using a c-Myc antibody to detect binding to DNA within the DLEU2 and SMC4 promoter regions. DNA enrichment in chromatin immunoprecipitated by the c-Myc antibody was determined by Real-Time PCR. Relative fold-enrichment was then determined by comparing the enrichment with c-Myc antibodies to the enrichment for the no antibody control (Relative enrichment = sample enrichment (Myc)/sample enrichment (no antibody)). A negative control for Myc binding was included that corresponded to a region in chromosome 1 (chr1:204,366,822-204,366,872). Data are mean ±SEM from 3 independent measurements.