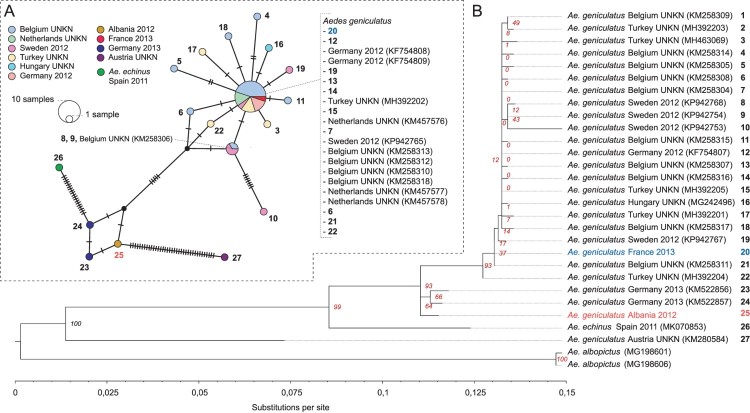
Figure 1.
Cytochrome oxidase I gene (COI) sequence variation at the intraspecific level among European Ae. geniculatus mosquitoes. A – Haplotype network inferred by the TCS method using a 437 bp mitochondrial DNA sequence (COI gene) covered by 39 specimens identified as Ae. geniculatus or Ae. echinus. The size of the each circle represents the frequencies of the haplotype, with each colour showing the geographical origin and collection date of the specimen. The term UNKN was display when the collection date was not known. Mutations are shown as perpendicular bars along the branches and black small circles represent inferred unsampled haplotypes. Haplotypes in common with the maximum-likelihood phylogenetic tree (B) are represented with numbers. B – Phylogenetic relationships between unique COI haplotypes from Ae. geniculatus, Ae. echinus and Ae. albopictus mosquitoes. Inferences were calculated with a maximum-likelihood method implemented in RAxML v.8.2.10 [30]. Topology was rooted to the Ae. albopictus (MG198601) COI sequence. Bootstrap support values are indicated on each node in red. COI sequences from single Ae. geniculatus specimens from Tirana, Albania (mosquito population that was used in this study) and Paris, France are represented in red and blue, respectively. GenBank accession numbers are displayed for each sequence.