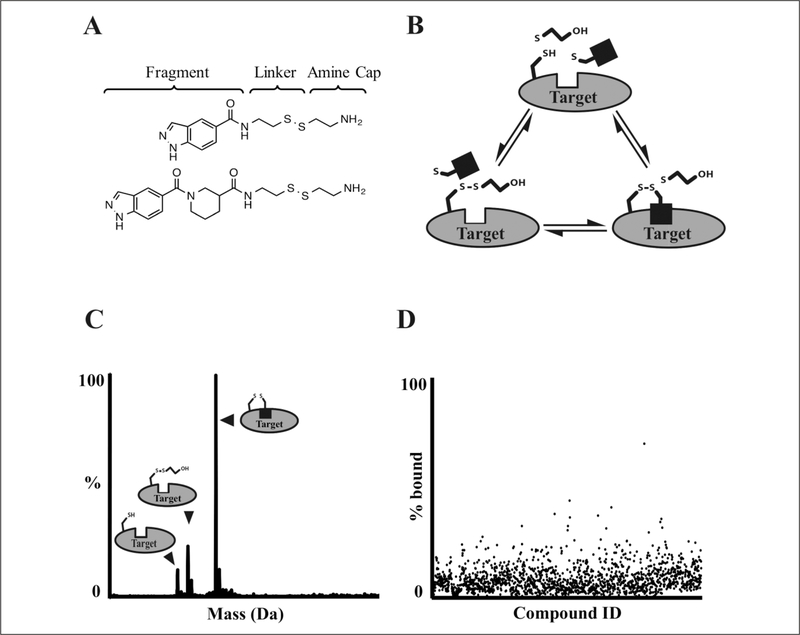
Figure 1.
Liquid chromatography/mass spectrometry (LC/MS) screening workflow. (A) Examples of structures from the tethering library.15,16 (B) Labeling reaction scheme. Target protein, β-mercaptoethanol (βME), and various fragments (black square) are mixed in individual wells of a 384-well plate and incubated until equilibrium is reached. (C) Rapid ultra-pressure liquid chromatography (UPLC) desalting, time-of-flight (TOF) detection, and m/z deconvolution identify unlabeled, βME-capped, and fragment-bound protein species. (D) Detected species are checked for expected fragment adduct formation and plotted as a percentage of protein that is fragment bound. Results are checked for data quality and uploaded to an internal database where hits are selected for follow-up.