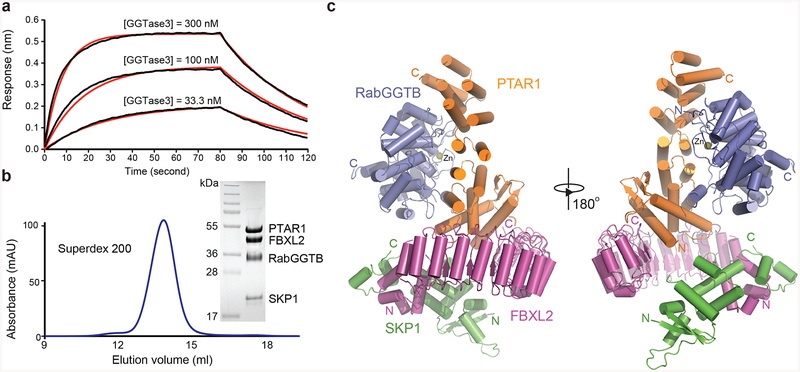
Figure 3. Overall structure of GGTase3-FBXL2-SKP1 complex.
(a) GGTase3 binding to FBXL2-SKP1 monitored by Octet BioLayer Interferometry analysis in the absence of GGPP or its analog. Sensorgram traces (black lines) of the interactions of His-FBXL2-SKP1 with GGTase3 at three concentrations (300 nM, 100 nM, and 33.3 nM) overlaid with curve fits (red lines). The global fit yields a calculated KD of 69.0 nM ± 1.1 nM.
(b) Size exclusion chromatography analysis of the purified GGTase3-FBXL2-SKP1 protein complex with sodium dodecylsulfate-polyacrylamide gel electrophoresis analysis of the central fraction containing the protein complex with all four subunits in equal stoichiometry.
(c) Two orthogonal views of the tetrameric complex containing PTAR1 (orange), RabGGTB (slate), FBXL2 (magenta) and SKP1 (green). The zinc ion at the active site of RabGGTB is shown in yellow sphere. The N and C termini of different proteins are labeled N and C in corresponding colors.