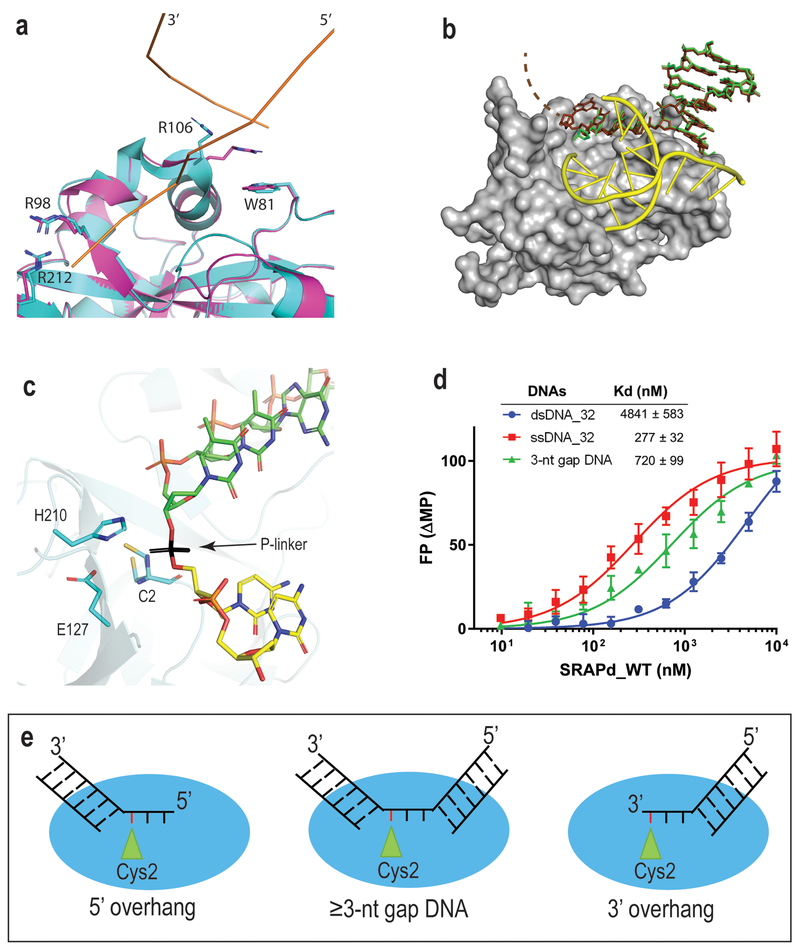
Figure 2. SRAPd interaction with Potential DNA-damage repair substrates.
(a) Crystal structure of Apo_SRAPd in magenta, superposed with SRAPd_3nt in cyan, at the dsDNA-interaction site A. DNA-A is shown as ribbon representation in orange. (b) Crystal structure of SRAPd_6nt in complex with six-nucleotide 3’ overhang. SRAPd is shown in surface representation in grey. The six-nucleotide overhang at the dsDNA-interaction site A is shown as a stick model in brown superposed with SRAPd_3nt three-nucleotide 3’ overhang, in green. The electron density for the last two nucleotides in SRAPd_6nt was not resolved and not modeled (indicated by dashed lines). DNA-B at the dsDNA-interaction site B is shown in yellow. (c) The 3’ and 5’ ends of two DNA molecules at the catalytic triad site of SRAPd_3nt are in close enough proximity to be linked by a phosphate group, shown in black color for the purposes of illustration. (d) Fluorescence polarization DNA-binding affinities of HMCES SRAPd to ssDNA, dsDNA, and dsDNA containing a three-nucleotide gap (3-nt gap DNA). Experiments were performed in triplicates and data are represented as mean +/− SD. (e) A model illustrating the potential DNA damage substrates that can be recognized by HMCES. Red line represents the abasic site in DNA. Source Data for panel d are available online.