Extended Data Fig. 9. Single cell RNA-Seq analysis reveals unique features associated with memory B cells recruited to the FRT.
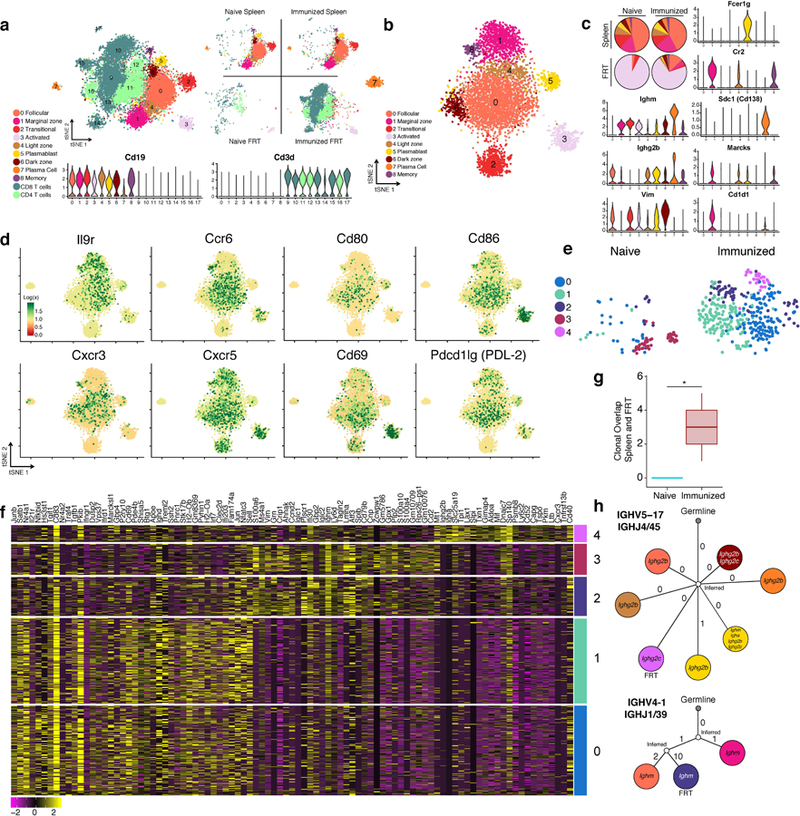
Spleens and FRT of C57BL/6 mice were collected for 10x single cell RNA-Seq one day after (re-)challenge with HSV-2; spleens were sorted for B cells and FRT were sorted for B and T cells (n= 8 pooled mice). a, tSNE plot associated with 5’ gene expression profiling of 13,763 cells captured on the 10x Chromium platform. B cell identities were determined by re-clustering CD19+ populations after initial clustering and reassigning them according to analysis in b, refer to methods for more detail. Violin plots depict kernel density estimates to show the distribution of expression values and was generated using the default VlnPlot function in Seurat (n = 13,763 cells). b, tSNE plot associated with CD19+ clusters identified from a after re-clustering (n = 5,390 cells). c, Relative cell population frequency in the different compartments and conditions are shown in pie charts and gene expression of each cluster is shown in violin plots. Violin plots depict kernel density estimates to show the distribution of expression values and was generated using the default VlnPlot function in Seurat (n = 5,390 cells). d, Heatmaps of genes in B cells associated with activation and chemokine/cytokine receptors (n = 5,390 cells). e, tSNE plots of B cells found in the FRT of naïve infected and immunized re-challenged mice after re-clustering only FRT B cells (n = 517 cells). f, Heatmap of top genes differentially expressed in each cluster in e. g. Clonal overlap between spleen and FRT of mice after primary or secondary challenge was measured using single cell BCR sequencing data, confidence intervals for clonal overlap were computed by bootstrap analysis and significance was assessed for non-zero overlap (p=0.003 by clone, p=0.0005 by sequence). Clonal overlap was computed from all productive BCR sequences from the 5,390 cells. Box plots depict number of clonal overlap calculated from bootstrap analysis with whiskers depicting upper percentile of 95th and lower percentile of 5th and box depicting 80th and 20th percentile. h. Lineage tree of clones that were shared in the spleen and FRT compartment, colors of cell types for splenic B cells and FRT B cells correspond to legends in c and e respectively.
