Fig. 1.
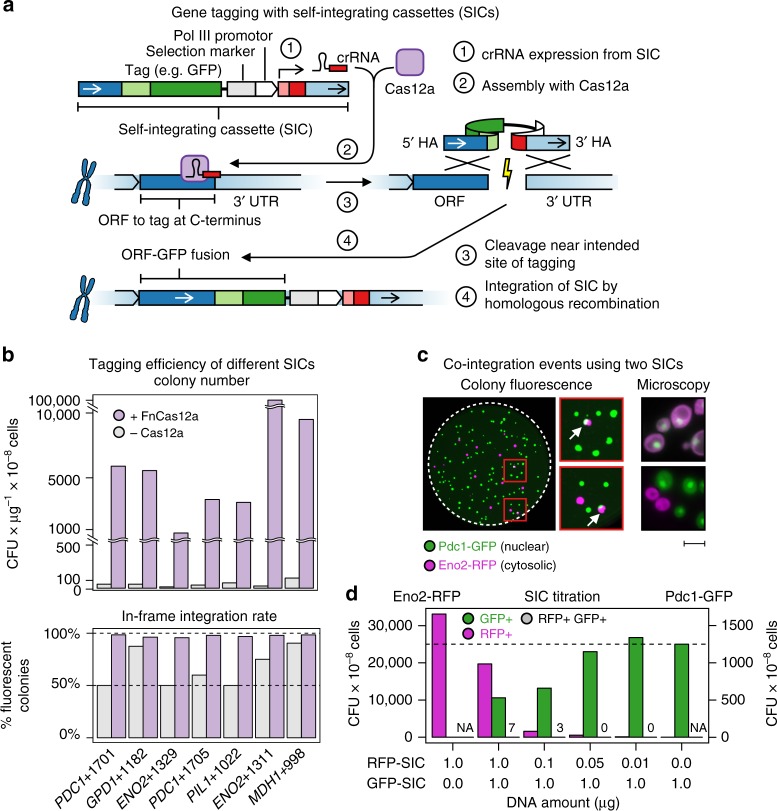
CRISPR-Cas12a-assisted single gene-tagging in yeast. a After transformation of the self-integrating cassette (SIC) into a cell, the CRISPR RNAs (crRNA) expressed from the SIC directs a CRISPR-Cas12a endonuclease to the genomic target locus where the DNA double strand is cleaved. The lesion is repaired by homologous recombination using the SIC as repair template so that an in-frame gene fusion is observed. b Efficiency of seven SICs of C-terminal tagging of highly expressed open-reading frames (ORFs) with a fluorescent protein reporter, in the absence (gray) or presence (purple) of Francisella novicida U112 (FnCas12a). Colony-forming units (CFUs) per microgram of DNA and cells used for transformation, and integration fidelity by colony fluorescence are shown. c Co-integration events upon simultaneous transformation of two SICs directed against either ENO2 or PDC1. Both SICs confer resistance to Geneticin (G-418), but contain different fluorescent protein tags. Colonies exhibiting green and red fluorescence (arrows) were streaked to identify true co-integrands. False-color fluorescence microscopy images show nuclear Pdc1-GFP (green fluorescent protein) in green and the cytosolic Eno2-RFP in magenta; scale bar 5 µm. d Titration of both SICs against each other (lower panel) with evaluation of GFP-tagged (GFP+), red fluorescent protein (RFP)-tagged (RFP+) or co-transformed (GFP+ RFP+) colonies. b–d Source data are provided as a Source Data file