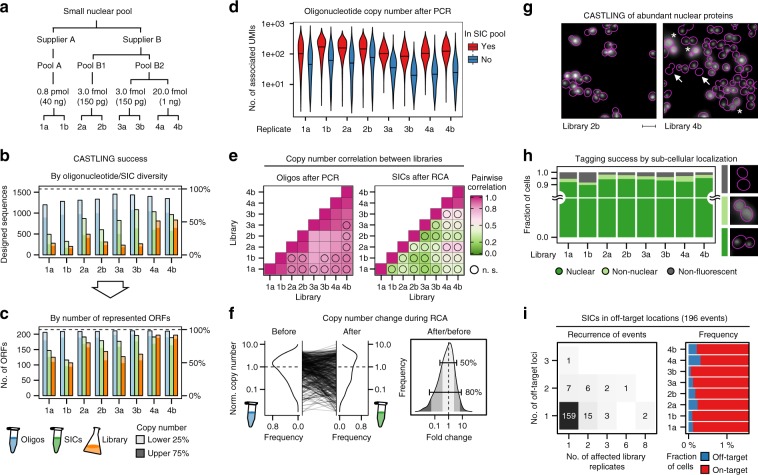
Fig. 3.
CRISPR-Cas12a (Cpf1)-assisted tag library engineering (CASTLING) for tagging 215 nuclear proteins with a green fluorescent protein. a Three oligonucleotide pools of the same design (1577 sequences, Supplementary Table 1) were used to create four tag libraries by CASTLING in duplicate sampling the indicated amount of starting material for PCR. b Detected oligonucleotide sequences of the design after PCR amplification (blue), self-integrating cassette (SIC) assembly (green), and in the final library (orange); oligonucleotides with copy number estimates (unique molecular identifier (UMI) counts) in the lowest quartile (lower 25%) are shown in light shade. c Same as b, but evaluated in terms of open-reading frames (ORFs) represented by the oligonucleotides or SICs. d Copy number of PCR amplicons recovered (red) or lost (blue) after recombineering; black horizontal lines indicate median UMI counts. e Pearson’s pairwise correlation of oligonucleotide or SIC copy number between replicates after PCR or rolling circle amplification (RCA), respectively; n.s., not significant (p > 0.05). f Kernel density estimates of copy number in replicate 1a as normalized to the median copy number observed in the oligonucleotide pool (before recombineering) and after recombineering into the SIC pool (left panel); the distribution of fold changes (right panel) highlights two frequency ranges: [0.1–0.9], that is, 80% of SICs, and [0.25–0.75], that is, 50% of SICs. g Representative fluorescence microscopy images of cells displaying nuclear, diffuse non-nuclear (asterisks), or no mNeonGreen fluorescence (arrows); scale bar 5 µm. h Quantification of fluorescence localization in >1000 cells in each replicate. i Recurrence of off-target events as revealed by Anchor-Seq across all library replicates and all genomic loci (left panel); the fraction of cells with SICs integrated at off-target sites (blue) within each clone population (red) is shown (right panel, axis trimmed). b–i Source data are provided as a Source Data file