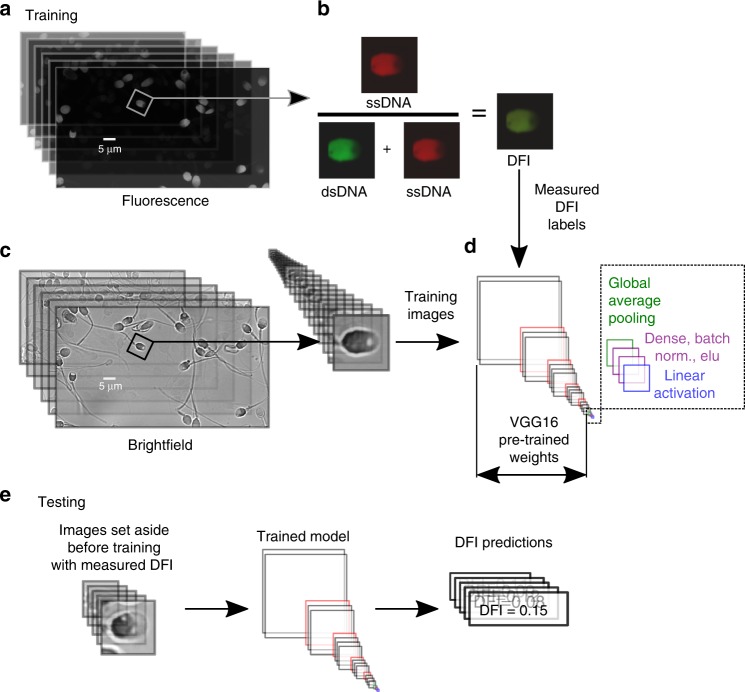
Fig. 1.
Experimental and modeling schematic. We illustrate the extraction of individual (a) fluorescence images to calculate the (b) DNA fragmentation index (DFI), as well as extraction of sperm head image from (c) bright-field microscopy images, which were used to train the (d) deep convolutional neural network. DFI was found using the acridine orange (AO) test39,37 (with brief details given in Methods and full details in Wang et al.38) and calculated as the ratio of red fluorescence (from presence of single-stranded DNA, ssDNA) to the sum of red and green fluorescence (from double-stranded DNA, dsDNA). The bright-field image was then labeled with the DFI value to train the model. The VGG16 network was modified by appending global average pooling and two dense layers with batch normalization and exponential linear unit (ELU) activation functions, after which linear activation was applied to condense the result to a single scalar value (DFI). e Once the model was trained, we fed images not used in training (but with measured DFI) and predicted the DFI, thereby yielding the generalizability of the model to unseen images