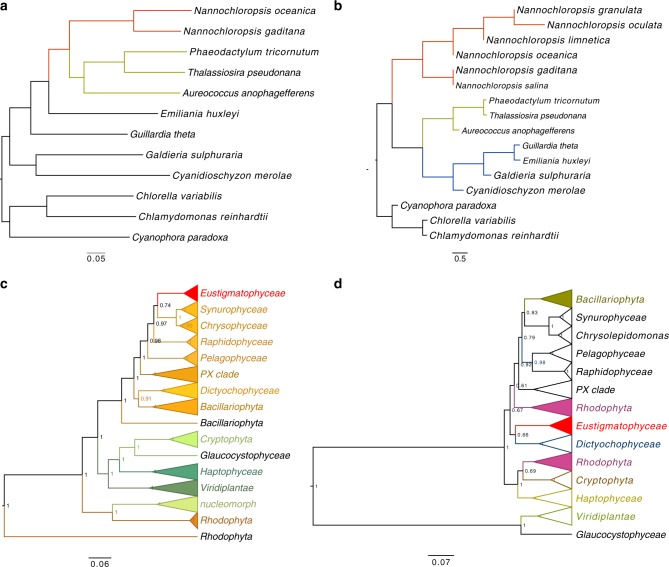
Fig. 2.
Phylogeny of Nannochloropsis and their relatives in different taxa deduced from nuclear genome (a), chloroplast genome (b), 18S ribosomal RNA gene (18S rDNA, c) and ribulose bisphosphate carboxylase large chain gene (rbcL) (d). a The phylogenetic tree of species inferred from the OrthoFinder using the genome protein sequences each species from the whole genome. b The phylogenetic tree of species inferred from the OrthoFinder using the chloroplast genome sequences each species. c The phylogenetic trees for 18S rDNA. The tree shows the consensus tree topology inferred by Bayesian analysis using alignments of 18S genes from NCBI. The scale bar indicates the nucleotide substitutions per site. This consensus topology derived from 512 trees, lnL = 22033.73. d The phylogenetic trees for rbcL protein. The tree shows the consensus tree topologies inferred by Bayesian analysis using alignments of rbcL proteins from NCBI. Scale bars represent 0.1 amino acid substitutions per site. In total, 439 aligned amino acid sites were analyzed. This consensus topology derived from 726 trees, α = 0.47 (0.41 < α < 0.56), pI = 0.0019 (0.0000007 < pI < 0.0059) and lnL = 10364.8