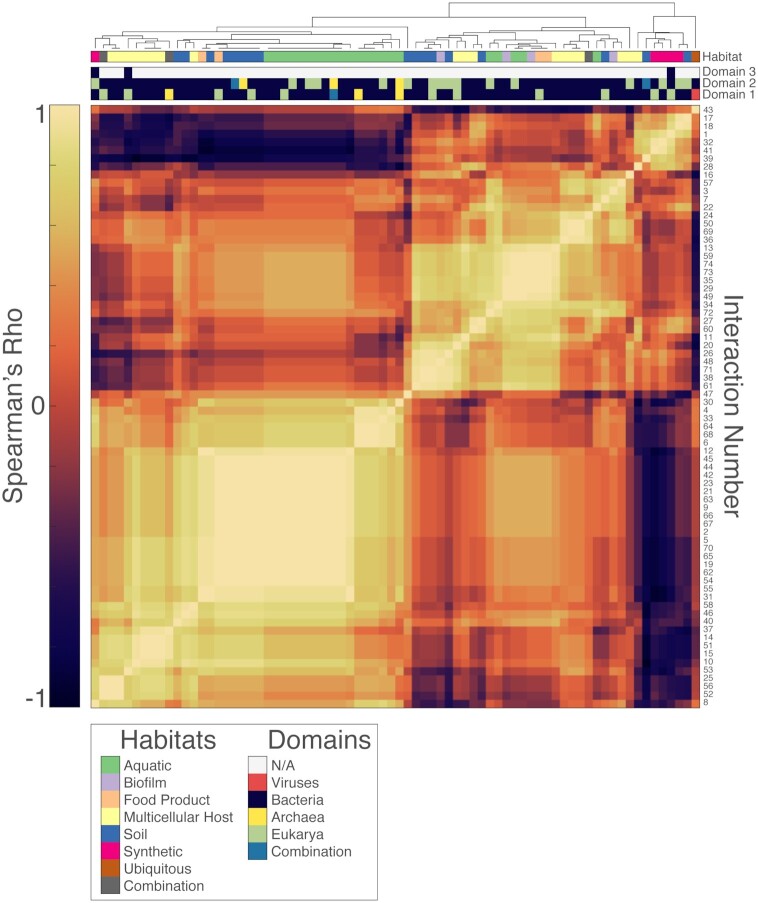
Figure 2.
Hierarchical clustering of microbial interactions, numbered according to their catalog entry in Table S1 (Supporting Information). Numerical values for interaction attributes (specificity, costs, ecological outcomes, dependencies, site, habitat and compounds involved) were normalized from 0 to 1. Multi-column attributes (i.e. those that contain specific values for individual participants, such as ecological outcome and cost) were additionally encoded into single unique values using the Cantor pairing function. Unknown values (comprising 2.1% of the dataset) were imputed with the mean of each column to enable all interactions to be compared. The normalized values were used to calculate pairwise distances between each interaction using Spearman's rho. Hierarchical clustering was then performed to generate a tree based on the resulting distance matrix. Taxonomic information (which was not used to generate clustering), as well as habitat information is displayed for all interactions.