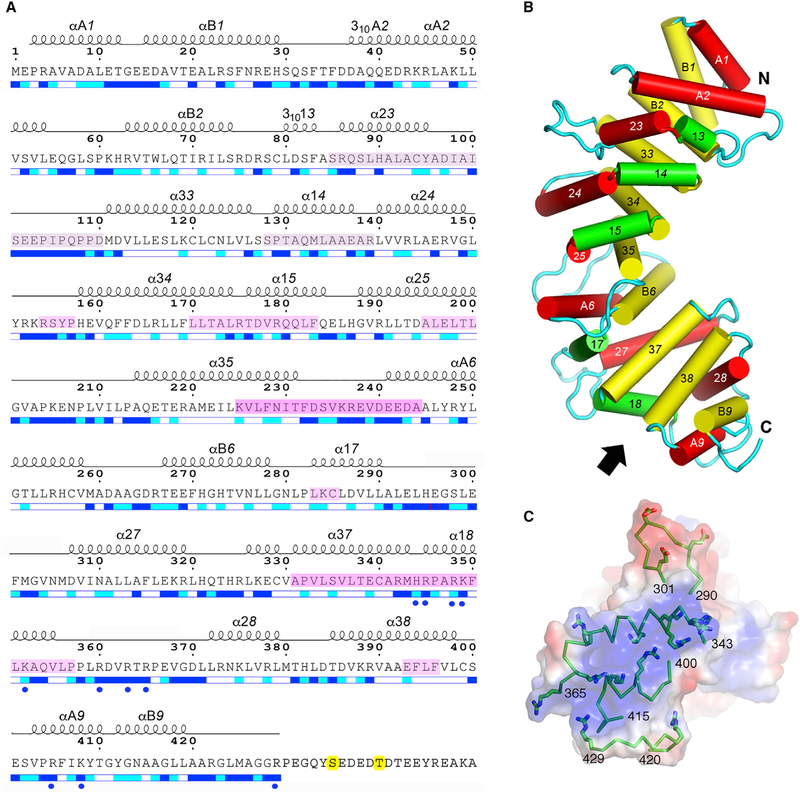
Figure 2. Annotated Amino Acid Sequence and Structural Features of pR452.
(A) Amino acid sequence of R452 showing the numbering convention for structural repeats. ARM repeats consist of three helices: α1X, α2X, and α3X, where X is the nth repeat of the structure. HEAT repeats consist of two helices: αAX and αBX. α-Helical secondary structure is shown as a series of loops above the amino acid sequence. Helices with 310 hydrogen bonding and geometry are so labeled. Straight-line sections indicate loop segments. No electron density is observed beyond residue 423 (molecule A) and 429 (molecule B). Magenta-tinted overlay on segments of the amino acid sequence indicate regions that are protected from hydrogen-deuterium exchange by Gαi1 (Kant et al., 2016), with color intensity proportional to degree of protection as shown in Figure 4. The two phosphorylation sites, Ser435 and Thr440, are highlighted in yellow. Blue and cyan bars shown below the amino acid sequence indicate residues that are solvent accessible (blue), partially accessible (cyan), or buried (white). Figure modified from output from ESPript 3.0 server (Robert and Gouet, 2014). See also Figures S3 and S4.
(B) Schematic representation of the structure of pR452. Cylinders represent a helices. Rendered in red are the ‘‘A’’ helices of HEAT repeats or the second helix of an ARM triad. ‘‘B’’ helices of HEAT repeats or the third helix of an ARM repeat are colored yellow, and the first helices of ARM repeats are rendered in green. See also Figure S4.
(C) Electrostatic potential (positive, blue; negative, red) at the molecular surface of the C terminus of pR452, viewed from the direction indicated by the arrow in (B), showing secondary structure (ribbons) and side chains of positively charged residues (stick figures) of residues 343–365 (α18) and residues 400–415 (αA9, αB9). Molecular surface and electrostatic potential computed using PyMOL Molecular Graphics System Version1.7 Schrödinger LLC.