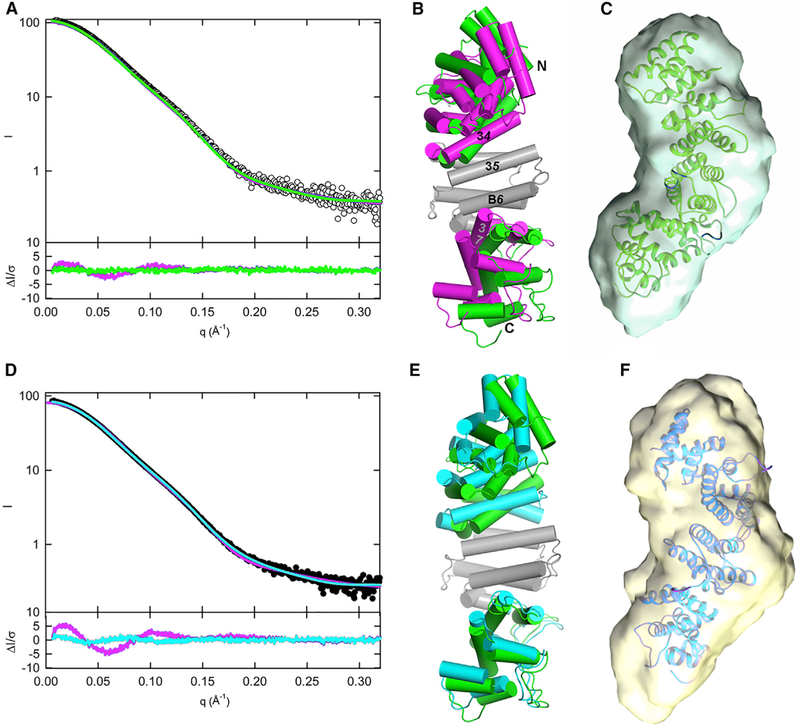
Figure 4. Solution Structures of pR452 and R452 Modeled from SAXS.
(A and D) Scattering profiles for pR452 (A, open circles) and R452 (D, closed circles). Computed scattering profiles from the crystallographic coordinates of pR452 and R452 are shown in magenta. Solution scattering profiles generated from normal mode refinement of pR452 crystallographic coordinates with respect to experimental scattering curves are shown for pR452 (A, green), and R452 (D, cyan). The bottom panels of (A) and (D) show the error-weighted residual difference plots ΔI/σ = [Iexperimental(q)– CImodel(q)]/σ(q) versus q, where C is a normalizing scale factor. Normal mode refinement of pR452 coordinates affords improved fits to the experimental scattering curves.
(B) Superposition of the crystal structure pR452 (magenta) onto the model generated by normal mode refinement of the latter with respect to the solution scattering curve for pR452 (green). Models, rendered as cylinders for α-helical segment. Helices at subdomain interfaces and the N and C termini are labeled. The coordinate sets were superimposed using the Cα atomic positions of the central rigid body domain, residues 184–287. See also Table S2.
(E) Superposition of the modeled solution structure of pR452 (green), as shown in (B), onto the modeled solution structure of R452 (cyan).
(C and F) Solution structure models of pR452 (green) and R452 (cyan) fit to their respective ab initio molecular envelopes. See also Figure S8.