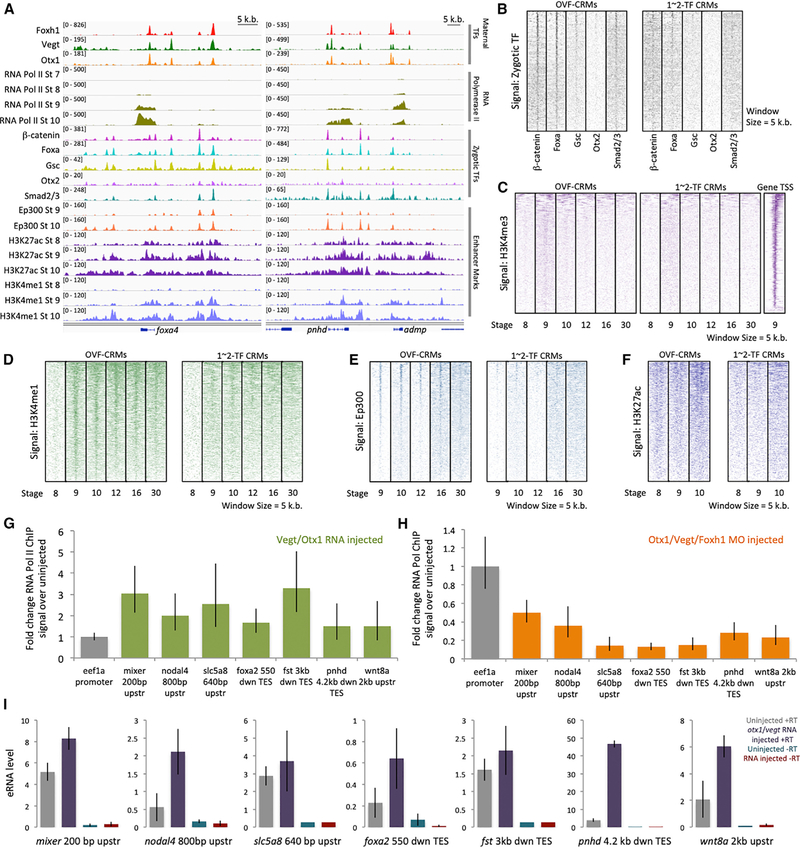
Figure 5. Combinatorial Otx1, Vegt, and Foxh1 Binding Pre-marks Zygotic cis-Regulatory Modules.
(A) Genome browser of maternal Otx1, Vegt, and Foxh1 binding with Pol II binding, mesendodermal zygotic TF binding (b-catenin, Foxa, Gsc, Otx2, and Smad2/3), and enhancer marks (Ep300, H3K27ac, and H3K4me1) near the genes foxa4, pnhd, and admp.
(B) Zygotically active TF ChIP-seq signal of b-catenin, Foxa, Gsc, Otx2, and Smad2/3 in OVF-CRMs and 1- to 2-TF CRMs during the gastrula stage.
(C) Heatmap of H3K4me3 promoter mark ChIP-seq signal in OVF-CRMs, 1–2 TF CRMs, and gene promoters as a positive control.
(D–F) Heatmap of enhancer marks H3K4me1 (D), Ep300 (E), and H3K27ac (F) ChIP-seq signal in OVF-CRMs and 1-to 2-TF CRMs.
(G) Fold change of Pol II ChIP-qPCR signal at gastrula stage 10.5 from animal caps ectopically expressing otx1 and vegt mRNA assaying for association with OVF-CRMs, compared to uninjected animal caps. Shown is a single representative biological replicate. The error bars represent the variation from 3 technical replicates.
(H) Fold change of Pol II ChIP-qPCR signal at stage 10.5 from vegetal masses in Otx1, Vegt, and Foxh1 triple morpholino KD conditions assaying for association with OVF-CRMs, compared to uninjected vegetal masses. Shown is a single representative biological replicate. The error bars represent the variation from 3 technical replicates.
(I) RT-qPCR assay for eRNA transcription from OVF-CRMs in uninjected, and otx1 and vegt mRNA injected animal caps at stage 10.5. Genomic DNA contamination is quantified using an RT-qPCR control reaction with no reverse transcriptase (–RT). Shown is a single representative biological replicate. The error bars represent the variation from 3 technical replicates.
The experiments for (G)–(I) were repeated at least twice.