Abstract
Epstein-Barr virus (EBV) infection is ubiquitous worldwide and is associated with multiple cancers, including nasopharyngeal carcinoma (NPC). The importance of EBV viral genomic variation in NPC development and its striking epidemic in southern China has been poorly explored. Through large-scale genome sequencing of 270 EBV isolates and two-stage association study of EBV isolates from China, we identified two non-synonymous EBV variants within BALF2 strongly associated with the risk of NPC (odds ratio (OR) = 8.69, P=9.69×10−25 for SNP 162476_C; OR = 6.14, P=2.40×10−32 for SNP 163364_T). The cumulative effects of these variants contributed to 83% of the overall risk of NPC in southern China. Phylogenetic analysis of the risk variants revealed a unique origin in Asia, followed by clonal expansion in NPC-endemic regions. Our results provide novel insights into NPC endemic in southern China and also enable the identification of high-risk individuals for NPC prevention.
Editorial Summary
Whole-genome sequencing and association analysis of 270 Epstein-Barr virus (EBV) isolates from China identify two non-synonymous EBV variants within BALF2 strongly associated with the risk of nasopharyngeal carcinoma.
Epstein-Barr virus (EBV) was discovered in 19641,2 and is the first human virus to be associated with cancers, including nasopharyngeal carcinoma (NPC), a subset of gastric carcinoma, and several kinds of lymphomas3. Although EBV infection is ubiquitous in human populations worldwide, its most closely associated malignancy, NPC, has a unique geographic distribution. Rare in most of the world, NPC is a very common cancer in southern China, where the incidence rate can reach 20 to 40 cases per 100,000 individuals per year4. Multiple human susceptibility loci, including HLA, CDKN2A/2B, TNFRSF19, MECOM, and TERT loci, have been discovered for NPC, but the contributions of these loci to overall risk are limited5–8. Moreover, the risk variants at these loci are widely distributed in the Chinese population and therefore cannot explain the unique endemic of NPC in southern China. Thus, the cause of NPC, commonly known as the Cantonese cancer, remains unknown.
Since the first EBV genome sequence, B95–8, was published in 19849, more than 100 EBV genomes have been sequenced in spontaneous lymphoblastoid cell lines and patients with EBV-associated diseases. These studies revealed important genomic variations among EBV isolates from different geographic origins10–15. Although the importance of EBV genome variation in the risk of EBV-associated diseases has been explored15–18, these studies suffered from the confounding effect of geographic distribution and insufficient sample size. As a result, robust epidemiological and genetic evidence linking specific EBV strains to the pathogenesis of NPC is lacking.
In the current study, we performed large-scale whole-genome sequencing (WGS) of 215 EBV isolates from patients diagnosed with EBV-associated cancers (including NPC, gastric carcinoma, and lymphomas) and 54 isolates from healthy controls recruited from both NPC-endemic and non-endemic regions of China. Through a comprehensive and systematic association analysis of EBV genomic variation and subsequent replication analysis in an independent sample, we identified two non-synonymous variants in the BALF2 gene associated with high risk for NPC. These two variants explain 83% of the overall risk in NPC-endemic southern China. In addition, phylogenetic analysis of EBV isolates from the current study and worldwide strains suggest a unique Asian origin followed by a clonal expansion of the two NPC-high-risk variants in southern China. Thus, we have discovered the high-risk EBV subtypes that contribute significantly to the overall risk of NPC, as well as its unique epidemic in southern China.
Results
EBV whole-genome sequencing
Using a capture-based protocol, we obtained EBV genome sequences from 215 samples of tumor, saliva, and plasma from EBV-associated cancer patients (NPC, gastric carcinoma, and lymphomas) and 54 saliva samples from healthy donors, as well as one genome from NPC cell line C666–1 (For an overview of the study, see Supplementary Fig. 1, Supplementary Tables 1–4 and Methods). Of the 270 EBV isolates, 221 were obtained from the NPC-endemic region of southern China (Guangdong and Guangxi Provinces), and 49 were from NPC-non-endemic regions of China. The average sequencing depth of all the isolates was 1,282×, and on average, 95% of the EBV genome was covered with at least 10× coverage (Supplementary Fig. 2). Using B95–8 as the reference, we identified a total of 8,469 variants (8,015 SNPs, 454 INDELs) across the EBV genome (for variant statistics, see Supplementary Table 5 and Supplementary Fig. 2). The number of variants identified in each sample ranged from 1,006 to 2,104, with EBNA-2, −3A, −3B and −3C and LMP-2A and −2B being the most polymorphic genes (Supplementary Fig. 3), consistent with other reports14–16. To explore the accuracy in sequencing and variant calling, we compared the re-sequenced C666–1 EBV genome against the published record and found a high concordance rate of 97.9%19 (Supplementary Table 6). In addition, when subsets of variants discovered by EBV whole genome sequencing (WGS) were re-genotyped by Sanger sequencing and MassArray iPLEX assay, 97.55% and 99.99% of tested variants were confirmed, respectively (Supplementary Tables 7 and 8). Both results indicate that our sequencing and variant calling procedures were highly accurate.
To understand intra-host polymorphism within an individual, two EBV fragments were amplified and sequenced in paired saliva and tumor samples from 25 patients with NPC. The variant difference between the paired saliva and tumor samples (median 1.1%, 1st to 3rd quartile: 0–3.4%) was substantially lower than the between-host difference (median 13.5%, 1st to 3rd quartile: 3.7–16.9%) (Supplementary Fig. 4). In addition, we sequenced the EBV whole genomes from the same NPC patient in paired tumor and saliva samples and observed a 99.27% concordance between the variants in EBV tumor and saliva isolates (Supplementary Table 9). Taken together, these observations suggest that paired saliva and tumor samples from the same subject had the same EBV genome or strain. Therefore, we combined the genome sequence information from tumor and saliva samples from NPC cases in subsequent analyses.
BALF2 gene region showing strongest association
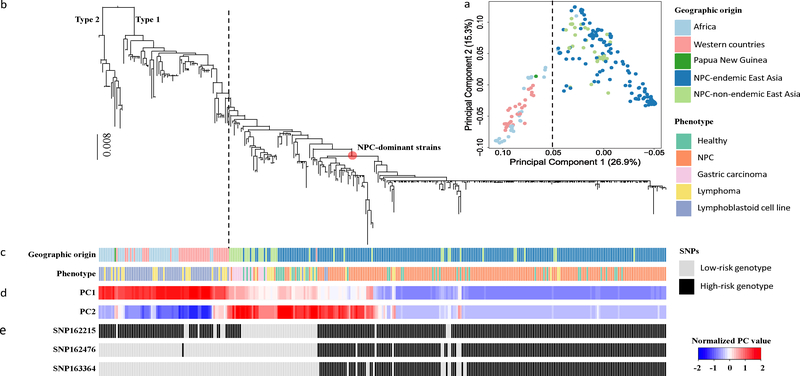
To investigate the impact of EBV genomic variations on NPC risk, we performed a two-stage genome-wide association study. In the Discovery phase, we included the EBV genomes from 156 NPC cases and 47 controls from the 270 EBV-WGS isolates. These isolates included in the discovery phase are exclusively from Guangdong and Guangxi Provinces in the NPC-endemic region of southern China. A principal component analysis (PCA) of the human genome variation of all the cases and controls with the reference population samples from the 1000G project20 confirmed their ethnic origin and the genetic match between cases and controls (Supplementary Fig. 5). We also performed PCA analysis of EBV genomes using all the 270 strains from the current study together with 97 publicly available genomes. The distribution of the EBV strains along the first principal component (PC) was continuous, ranging from Africa and Europe to Asia (Fig. 1a). Within Asia, the second PC showed a partial separation of the isolates from NPC-endemic region and non-endemic region of Asia (Fig. 1a, d).
Figure 1. Principal component and phylogenetic analyses of EBV genomes.
(a) Principal component analysis of 270 EBV isolates sequenced in the current study and 97 published isolates. The first two principal-component scores (PC1 and PC2) are plotted. Explaining 26.9% of the total genomic variance, PC1 discriminates between East Asian and Western/African strains; PC2 explains 15.3% of the total variance. (b) Phylogeny of 230 EBV single strains sequenced in the current study and 97 published strains. Macacine herpesvirus 4 genome sequence (NC_006146) was used as the outgroup to root the tree. Type 1 and Type 2 EBV lineages are indicated. The red dot on the phylogeny indicates the lineage of the NPC-dominant EBV strains, where 22 of 37 strains from healthy controls in NPC-endemic regions in southern China were located. Dashed lines in (a) and (b) indicate the separation between East Asian and Western/African strains. (c) Geographical origins and phenotypes of samples from which EBV strains were sequenced are shown with colors as indicated. (d) The normalized values of PC1 and PC2 scores are shown from blue to red. (e) The genotypes of SNPs 162215C>A, 162476T>C, and 163364C>T in each isolate.
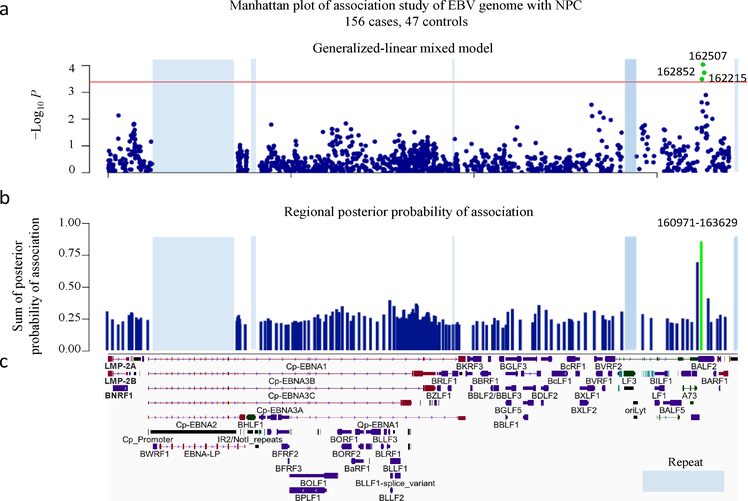
To control for the potential impact of the population structures of both the human and EBV genomes, the genome-wide association analysis was performed using a generalized-linear mixed model, with age, sex, the first four human PCs and previously reported NPC human GWAS SNPs (rs2860580 and rs2894207 at HLA locus, Supplementary Table 10, see Methods) as fixed effects and the genetic relatedness matrix of EBV genomes as random effects21. The discovery analysis revealed multiple association signals along the EBV genome. The strongest association was in the BALF2 region (NC_007605.1:162507C>T, P = 9.17×10−5) without any indication of inflation due to genetic structure (genomic control inflation factor λGC = 1.03; Fig. 2a, Supplementary Table 11 and Supplementary Fig. 6). We also investigated the association evidence for the recently reported EBER2 variants by Hui et al18 in our discovery dataset. NC_007605.1:7048A>C, which was a leading variant for the reported associations at EBER2 region, showed significant association in our genome-wide analysis (P = 1.25×10−7), but the significance was largely abolished by controlling for population structure (P = 1.52×10−2; Supplementary Fig. 6).
Figure 2. Genome-wide association analysis of EBV variants in 156 NPC cases and 47 controls.
(a) Manhattan plot of genome-wide P values from the association analysis using a generalized-linear mixed model. The −log10-transformed P values (y axis) of 1545 variants in 156 NPC cases and 47 controls are presented according to their positions in the EBV genome. The minimum P value (SNP 162507C>T) is 9.17×10−5. The red line is the suggestive genome-wide significance P value threshold of 4.07×10−4. The three SNPs 162507C>T, 162852G>T and 162215C>A reaching genome-wide significance are labeled as green. (b) The regional plot of the posterior probabilities of association. The EBV genome was partitioned into overlapping 20-variant bins with 10-variant overlap between adjacent bins. The sum of the posterior probabilities for variants was assigned to each region. The one region from position 160971 to 163629 with strong evidence (> 0.85) for association with NPC risk is shown in green. (c) Schematic of EBV genes. Repetitive regions in EBV genomes are masked by light blue.
In addition, we also performed a multi-SNP genome-wide association analysis using Bayesian variable-selection regression by piMASS22, which provided consistent and strong evidence for the association in the BALF2 region (posterior probability = 0.86; Fig. 2b). When we evaluated the statistical significance of association using permutation test (see Methods), only the associations within the BALF2 region reached genome-wide significance (suggestive genome-wide significance, P < 4.07×10−4). Consistent with the extensive linkage disequilibrium (LD) in the EBV genome (Supplementary Fig. 7), conditioning on the genetic effects of the SNPs in the BALF2 region greatly reduced the extensive associations across the entire EBV genome (Supplementary Fig. 8).
Fine-mapping and validation of BALF2 variants
We performed a Bayesian fine-mapping analysis to prioritize potentially causal SNPs in the BALF2 gene region using PAINTOR and found that only the three non-synonymous coding variants (NC_007605.1:162215C>A, 162476T>C, and 163364C>T) were significantly associated (Supplementary Fig. 9 and Supplementary Table 12). We genotyped these variants in an independent sample of 483 NPC cases and 605 age- and sex-matched healthy controls (Validation phase; Supplementary Table 13). To reduce the potential impact of population stratification, all the cases and controls were recruited from the single NPC-endemic region, Zhaoqing County, in the Guangdong Province of China. All three BALF2 SNPs were significantly associated with NPC risk in the independent sample (P < 0.017, 0.05/3), consistent with the discovery phase results (Table 1). The meta-analysis of the combined discovery and validation samples confirmed the associations with the three SNPs of BALF2 with genome-wide significance according to both permutation analysis (162215_C, odds ratio (OR) = 7.60, P = 1.42×10−18; 162476_C, OR = 8.69, P = 9.69×10−25; and 163364_T, OR = 6.14, P = 2.40×10−32; Table 1). All the three SNPs showed significant LD (Supplementary Fig. 10), but conditional analysis revealed that the associations with SNPs 162215C>A and 162476T>C were correlated, whereas SNP 163364C>T showed an independent association that also reached genome-wide significance (Table 1).
Table 1.
The association of three non-synonymous SNPs in BALF2 gene and the risk for NPC
| SNP | High-risk genotype | Discovery |
Validation |
Combined |
Odds ratio | 95% CI |
P value
conditional on SNPs |
Annotation | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 156 cases | 47 controls | P value | 483 cases | 605 controls | P value | 639 cases | 652 controls | P value | 163364 | 162476 | |||||
| 162215C>A | C | 96.15% | 65.96% | 3.22×10−04 | 95.03% | 74.71% | 9.92×10−16 | 95.31% | 74.08% | 1.42×10−18 | 7.60 | 4.97–11.62 | 7.78×10−05 | 1.94×10−01 | BALF2, V700L |
| 162476T>C | C | 93.59% | 61.70% | 5.09×10−03 | 94.00% | 65.12% | 1.94×10−23 | 93.90% | 64.88% | 9.69×10−25 | 8.69 | 5.79–13.03 | 1.10×10−06 | BALF2, I613V | |
| 163364C>T | T | 88.46% | 48.94% | 7.95×10−03 | 83.85% | 45.45% | 6.92×10−32 | 84.98% | 45.71% | 2.40×10−32 | 6.14 | 4.59–8.22 | 4.84×10−11 | BALF2, V317M | |
The association of three EBV SNPs with NPC risk was tested in discovery and validation samples and with a meta-analysis of the combined discovery and validation samples. Frequencies of high-risk genotypes in discovery, validation and combined analyses are indicated. Odds ratios conferred by high-risk genotypes and the 95% confidence intervals (CI) were estimated from the meta-analysis of the combined discovery and validation phases. Conditional regression analyses were performed in combined samples, and P values of SNP associations in conditional analyses are listed.
We further explored the association between the haplotypes (strains) composed of SNPs 162215C>A, 162476T>C and 163364C>T and the risk of NPC. Taking the haplotype composed of the 3 low-risk variants (A-T-C) as a reference, we found no association for the haplotype carrying the high-risk variant for SNP 162215_C (haplotype C-T-C: OR = 1.12; P = 0.78), although the number of haplotypes for comparison was limited (Table 2 and Supplementary Table 14). Both the haplotypes carrying the high-risk variants of either all three SNPs or only SNPs 162215_C and 162476_C showed strong risk effect (haplotype C-C-T: OR = 11.71, P = 2.39×10−24; haplotype C-C-C: OR = 3.50, P = 1.22×10−5; Table 2 and Supplementary Table 14), but haplotype C-C-T showed significantly stronger effect than did the haplotype C-C-C (P = 2.07×10−10), clearly indicating the additional risk effect of SNP 163364_T. The haplotype analysis further confirmed that NPC risk is primarily associated with SNPs 162476_C and 163364_T and that the association with SNP 162215_C needs to be further evaluated. We also performed pair-wise interaction analysis showing no evidence for interaction between SNPs 162476T>C and 163364C>T (P = 0.93). Finally, multiple regression analysis yielded independent risk effects (OR) of 3.31 for SNP 162476_C and 3.35 for SNP 163364_T (Supplementary Table 15), which were consistent with the risk effect of the haplotype carrying the two high-risk variants (haplotype C-C-T: OR = 11.71; Table 2).
Table 2.
EBV haplotypes composed of SNPs 162215C>A, 162476T>C and 163364C>T and the risk for NPC
| EBV subtype (162215–162476–163364) | 639 cases |
652 controls |
Odds ratio* | 95% CI | P value | ||
|---|---|---|---|---|---|---|---|
| no. | % | no. | % | ||||
| L-L-L (A-T-C) | 25 | 3.91% | 171 | 26.23% | - | - | |
| H-H-H (C-C-T) | 539 | 84.35% | 293 | 44.94% | 11.71 | 7.44–19.26 | 2.39×10−24 |
| H-H-L (C-C-C) | 57 | 8.92% | 118 | 18.10% | 3.50 | 2.02–6.24 | 1.22×10−05 |
| H-L-L (C-T-C) | 13 | 2.03% | 65 | 9.97% | 1.12 | 0.47–2.50 | 7.83×10−01 |
| other subtypes | 5 | 0.78% | 5 | 0.77% | 4.26 | 0.80–19.63 | 6.71×10−02 |
Odds ratios of individual EBV subtypes and 95% confidence intervals (CI) were estimated with a logistic model by categorizing each subtype as a single variable and adjusting for age, sex, the status of single- or multiple-infection and human GWAS SNPs (rs2860580 and rs2894207) in the combined discovery and validation data sets. Subjects with EBV subtype A-T-C, a common low-risk subtype, were used as the reference category. H represents the high-risk genotype; L represents the low-risk genotype.
Given the well-known function of BALF2 as the single-stranded DNA binding protein, a core component of viral DNA replication machinery23–25, we also investigated oral EBV abundance and its association with different BALF2 haplotypes in the 533 NPC cases and 651 controls. The viral DNA load varied widely across the samples, and viral DNA abundance in saliva was significantly lower in patients than in controls (P = 4.2×10−13; Supplementary Fig. 11). In both cases and controls, we observed the consistent decrease in viral load among individuals infected by the high-risk subtypes (C-C-T or C-C-C), especially C-C-C (P = 0.056), compared to the low-risk (A-T-C) haplotype (Supplementary Fig. 12), but the differences were marginally significant (Supplementary Table 16).
The evolution of the high-risk subtypes
In China, the frequency of the two high-risk haplotypes (C-C-T and C-C-C) was very high in the NPC-endemic region (93.27% in NPC cases and 63.04% in controls), but much lower in non-endemic areas (55% in NPC cases, 14.29% in controls; Supplementary Table 17). Interestingly, the two risk haplotypes were absent or extremely rare in non-Asian individuals from Africa and western countries (Supplementary Table 17), suggesting an Asian origin of the EBV high-risk variants. To further explore the evolution of the EBV risk variants, we investigated the phylogenetic relationship among the EBV strains from the current study and from published sequences. By examining the frequency and distribution of heterozygous SNPs, we identified 230 EBV single-infection strains from the 270 WGS isolates (see Methods, Supplementary Fig. 13 and Supplementary Table 18). With these 230 EBV isolates from the current study and 97 publicly available genomes, we performed phylogenetic inference. The evolutionary relationship among all sequences was highly unbalanced, with a deep split between Type 1 and Type 2 EBV isolates (Fig. 1b). All Type 2 EBV isolates were geographically restricted to Africa, as previously observed14,15,26. The Type 1 EBV clade showed a continuous branching starting from Africa, Europe, and Asia, matching the overall distribution along the first PC in the PCA analysis (Fig. 1b–d). As in previous studies17,27, 97% of 230 EBV single strain were found to be China 1 subtype, and 2% were China 2 (defined by LMP-1 classification; Supplementary Fig. 14). Within the Asian group, isolates from NPC-non-endemic areas clustered toward the basal position of the lineage, similar to the pattern observed along the second PC in the PCA map (Fig. 1b–d). The most striking pattern in the phylogenetic relationship was a rapid radiation of NPC-dominant strains in the endemic population from southern China. EBV genomes from NPC patients appeared to have expanded recently from a common ancestor, and more than half (22 of 37) of healthy controls from this region were infected with NPC-dominant strains (Fig. 1b, c).
When mapping the three SNPs of BALF2 (SNPs 162215C>A, 162476T>C and 163364C>T) onto the phylogenetic tree of the EBV genomes, we observed that all the strains carrying the risk variants of SNPs 162476_C and 163364_T were within the Asian subclade, whereas the carriers of the risk variant of SNP 162215_C had a much broader distribution (Fig. 1b, e). Within the Asian subclades, the carriers of SNPs 162476_C and 163364_T were enriched in the strains from NPC patients (NPC-dominant strains). These results provided strong evidence for the Asian origin of SNPs 162476_C and 163364_T and were consistent with their high-risk effect on NPC. The distribution of these genotypes also suggested that SNP 162215_C was less likely to be a risk variant for NPC, and its association effect was due to the results of its LD with SNP 162476_C (LD R2 = 0.67).
Discussion
Because of the ubiquity of EBV infection, the determinants of the distinctive geographical distribution of NPC have long puzzled the scientific community. Using large-scale sequencing and functional analyses, we discovered two EBV coding SNPs 162476_C and 163364_T that, to date, are the strongest known risk factors for NPC. The more than 6-fold increase in NPC risk conferred by these two high-risk EBV variants is far greater than the effects of any other known risk factors for this disease, including human genetic variants (Table 1 and Supplementary Table 10). In particular, with a population frequency of 45% and an OR of 11.71 (95% confidence interval (CI): 7.44 – 19.26), the EBV haplotype C-T of the two SNPs is the dominant NPC risk factor, contributing 71% (95% CI: 64–77%) of the overall risk of NPC in the endemic population of southern China. The second risk haplotype, C-C, also contributed about 10% of the risk, such that the two high-risk EBV haplotypes jointly accounted for 83% (95% CI: 76–90%) of NPC risk in this population (Supplementary Table 19). In non-endemic regions of China, the frequency of these high-risk haplotypes is much lower (about 10%), but they still contribute about 50% of the NPC risk driven by the strong risk effect. The frequency of the two high-risk EBV subtypes was not associated with the risk of developing other EBV-related cancers in our study, suggesting that their oncogenic effects might be specific to NPC. However, this observation would benefit from further work since our study was only powered to explore NPC.
Mapping these two causal variants onto the phylogenetic tree of EBV genomes revealed a distinct subclade of EBV subtypes carrying the two high-risk variants in Asia. The carriers were found only in Asia, thereby indicating an Asian origin for these two risk variants. Most interestingly, the phylogenetic analysis suggests a clonal expansion of these unique high-risk EBV subtypes in southern China. This expansion is consistent with the current distribution of these subtypes in China, with a very high frequency in the NPC-endemic region (93.27% in NPC cases and 63.04% in controls), but much lower in the non-endemic areas (55% in NPC cases and 9.68% in other non-NPC samples; Supplementary Table 17). At this point, we do not know what kind of selective phenotypes have driven the clonal expansion. More studies are needed to understand this evolutionary process. Taken together, the strong risk effect, the confined geographic distribution, the clonal expansion, and the extremely enriched frequency of these high-risk variants in the NPC-endemic region strongly suggest that these two EBV risk variants are the driving factors of the unique epidemic of NPC in southern China.
Our findings provide novel biological insights in EBV-mediated NPC tumorigenesis. The two risk variants 162476_C and 163364_T encode amino acid alterations in BALF2, the EBV single-stranded DNA binding protein, which is an abundantly expressed early lytic protein and a core component of viral DNA replication machinery23–25. Studies have shown that antibodies against EBV early lytic antigens, including BALF2, were highly enriched in the antibody signature for NPC risk prediction28,29, and BALF2 is also a frequent target of EBV-induced cytotoxic T cell response30. Because of the essential role of BALF2 in EBV lytic DNA replication, these amino acid changes may influence the productive lytic cycle of EBV by alternating the function of BALF2. This is consistent with our observation as well as others31 that the oral EBV abundance is lower in the NPC cases than the controls. In addition, we also observed the trend of decrease in oral EBV DNA load associated with the EBV subtype carrying the high-risk BALF2 haplotype, although this association is only marginally significant with a huge variation of saliva viral load among individuals. As demonstrated by previous reports32, this large variation of viral load in saliva was mainly due to the fact that EBV in buccal epithelium sporadically undergoes periodic lytic cycle with a large variation among different time points within an individual. Given the moderate impact of the BALF2 haplotypes on the overall variation of viral load, a much larger number of samples will help to confirm the statistical difference of viral load among the carriers of EBV with different BALF2 haplotypes. Taken together, our results and others suggested that the regulation of EBV lytic cycle plays an important role in the development of NPC. More molecular and functional investigations are needed to investigate this hypothesis and understand how the high-risk EBV subtypes and variants promote NPC tumorigenesis.
The discovery of these high-risk EBV variants also has important implications for public health efforts to reduce the burden of NPC, particularly in the endemic region of southern China. Testing for these high-risk EBV variants enables the identification of high-risk individuals for targeted implementation of routine clinical monitoring to detect NPC early. Primary prevention by developing vaccines against high-NPC-risk EBV strains is expected to lead to great attenuation of the Cantonese Cancer in China.
Methods
Study participants and samples
Participants of the current study were enrolled through two recruitments. The first was a hospital-based study enrolling patients with EBV-related cancers, including NPC, Burkitt lymphoma, Hodgkin lymphoma, NK/T cell lymphoma, and gastric carcinoma, as well as healthy controls from the Sun Yat-sen university Cancer Center in Guangdong Province, the First Affiliated Hospital of Guangxi Medical College in Guangxi Province, and the Affiliated Hospital of the Qingdao University in Shandong Province of China. The geographical origin of the participants covers the NPC-endemic area of southern China (Guangdong and Guangxi Provinces, where NPC has highest incidence of 20–40/100,000 individuals per year) and non-endemic regions in China where NPC is rare. After measuring EBV DNA level, 170 samples of tumor, saliva, and plasma were selected from the first recruitment for EBV whole-genome sequencing (WGS).
The second recruitment was a population-based, NPC case-control study enrolling NPC cases and healthy control subjects from Zhaoqing County, Guangdong Province (an NPC-endemic region). Cases and controls were matched by age and sex. Saliva samples were collected from all the subjects. After measuring saliva EBV DNA load in the second study, 99 saliva samples from 53 cases and 46 controls were selected for EBV WGS. Written informed consent was obtained from each participant before undertaking any study-related procedures, and both studies were approved by institutional ethics committee of Sun Yat-sen University Cancer Center.
Detailed sample information, including the geographic origin of the 270 isolates used for WGS, is summarized in Supplementary Tables 1–4. For the discovery phase of the EBV whole-genome association study (GWAS) with NPC, we included 156 cases and 47 controls exclusively from the NPC-endemic region from the 270 EBV WGS isolates. For the validation phase, 990 NPC cases and 1105 healthy controls from the endemic population-based case-control study were used by genotyping GWAS candidate SNPs (For details, see Supplementary Note).
Sample processing
Saliva samples were collected into vials containing lysis buffer (50 mM Tris, pH 8.0, 50 mM EDTA, 50 mM sucrose, 100 mM NaCl, 1% SDS). Tumor specimens were obtained from biopsy samples collected during surgical treatment and confirmed by histopathological examination. All saliva, tumor, and plasma specimens were stored at −80 °C. DNA was extracted from the saliva using the Chemagic STAR workstation (Hamilton Robotics, Sweden) and from the tumor biopsy, plasma and NPC cell line C666–1 using the DNeasy blood and tissue kit (Qiagen).
EBV genome quantification, whole genome sequencing, and variant calling
Using real-time PCR targeting a DNA fragment at the BALF5 gene (5’ and 3’ primers, GGTCACAATCTCCACGCTGA and CAACGAGGCTGACCTGATCC), we measured the EBV DNA concentration in each DNA sample with qPCR standard curve. Samples with EBV DNA concentration higher than 2500 copies per microliter were selected for viral whole genome sequencing (detailed information see the Supplementary Note).
The EBV genomes were captured using the MyGenostics GenCap Target Enrichment Protocol (GenCap Enrichment, MyGenostics, USA). After capture enrichment, DNA libraries were prepared and sequenced using the Illumina HiSeq 2000 platform according to standard protocols (Illumina Inc., San Diego, CA, USA). After raw sequence processing and quality control, paired-end reads were aligned to the EBV B95–8 reference genome (NC_007605.1) using the Burrows-Wheeler Aligner (BWA, version 0.7.5a)33,34. The average sequencing depth was 1,282 (range, 32 to 6,629). High genome coverage (average, 98.02%; range, 94.44% to 99.91%) was achieved (Supplementary Fig. 2).
Following GATK’s best practice (version 3.2–2), an initial set of 8,469 variants was first called after base and variant recalibration35. To avoid inaccurate calling, we further filtered out variants that had low coverage (depth < 10×) or were in repetitive elements or within 5 bp of an indel; 7,962 variants were retained for subsequent EBV phylogenetic, principal component, and association analyses. The functional annotation of the EBV variants was performed using the SNPEff package according to the reference genome (NC_007605.1, NCBI annotation, Nov 2013)36. A complete description of the sequencing and variant calling is presented in the Supplementary Note. No outlier was detected among the EBV isolates sequenced based on sequencing and variant statistics in the current study (Supplementary Fig. 2).
To evaluate the accuracy of our sequencing and variant calling, subsets of EBV variants were validated using either the Sanger sequencing or MassAarray iPLEX assay (Agena Bioscience). Two independent technologies can provide orthogonal evaluations of the sequencing accuracy. We amplified 299 PCR fragments from 53 randomly selected EBV isolates and re-sequenced them using the Sanger sequencing. The SNPs called by WGS and by the Sanger sequencing were 97.55% concordant (Supplementary Table 7). Similarly, the variants called by WGS and by the MassArray iPLEX assay were 99.99% concordant when genotyping 37 variants in 239 samples (Supplementary Table 8). In addition, when comparing the re-sequenced C666–1 EBV genome against the publicly available sequence19, the concordance was 97.93% (Supplementary Table 6).
To understand viral genomes from multiple sample types from the same patient, two EBV fragments (position 80,089 to 80,875 and position 81,092 to 81,829) containing 89 SNPs were resequenced using the Sanger method from paired saliva and tumor samples from the same set of patients. Across 25 NPC patients with paired tumor and saliva samples, the pairwise difference (defined as the genotype discordance rate at the 89 SNPs) between the tumor samples of the 25 patients (inter-host difference) as well as between the paired tumor and saliva samples of the same patient (intra-host difference) were calculated and compared (Supplementary Fig. 4). The median inter-patient difference was 13.5% (1st to 3rd quartile: 3.7–16.9%), and the median intra-host difference was only 1.1% (1st to 3rd quartile: 0–3.4%). The high concordance between variants from saliva and from tumors suggests that EBV sequences from paired saliva and tumor samples from the same patient are highly similar.
Genotyping analysis of EBV and human genetic variants by MassArray iPLEX
To genotype the EBV variants in the 990 cases and 1105 controls from Zhaoqing, the customized primers and the protocol recommended by the Agena Bioscience MassArray iPLEX platform were used. A fixed position in the human albumin gene was used as a positive control. Because the genotyping success rate strongly correlates with the EBV DNA abundance (Supplementary Fig. 15), about half of the validation samples (483 of the cases and 605 of the controls) could be successfully genotyped for all the three GWAS candidate markers (i.e., SNPs 162215C>A, 162476T>C and 163364C>T). The slightly lower success rate in the cases is consistent with the fact that the EBV DNA abundance was lower in the saliva from patients than from controls. For detailed information, see Supplementary Note.
Seven previously reported human SNPs in HLA (rs2860580, rs2894207 and rs28421666), CDKN2A/2B (rs1412829), TNFRSF19 (rs9510787), TERT (rs31489) and MECOM (rs6774494) were genotyped using customized primers and following the protocol recommended by the Agena Bioscience MassArray iPLEX platform in the 990 cases and 1105 controls from Zhaoqing. A fixed position in the human albumin gene was used as a positive control. The genotyping completion rate for all seven human SNPs was > 95%. Associations with NPC were assessed with logistic regression under an additive model adjusted for sex and age.
Determining single versus multiple EBV infections
The EBV genome usually undergoes clonal expansion in NPC tumors and other malignancies37–39. During clonal expansion, the EBV genome is stable, the intra-host mutation rate is often low, and heterozygous variants, as a result of quasi-species evolution within a host, are not frequent12,19,40. On the contrary, EBV isolates from specimens with multiple infections will have a higher number of heterozygous variants. We plotted the percentage of heterozygous variants across all the 270 samples from the WGS analysis and observed that heterozygosity (defined as a percentage of heterozygous variants) across all the samples showed two different distributions, with low and high numbers of heterozygous variants. By fitting two curves to the lower and higher quantiles of the empirical distribution, we defined the reflection point (i.e. the intersection of the two distributions) as the cutoff value (Supplementary Fig. 13). Samples with the proportion of heterozygous variants lower than the cutoff value were identified as single-infection samples, whereas samples above this threshold were identified as multi-infection samples. For the validation cohort, samples with the homozygous calls at all the three EBV SNPs were regarded as a single EBV subtype defined by BALF2 haplotypes. For samples with infection by multiple EBV subtypes, haplotypes of the three SNPs were inferred by Beagle 4.141. For details, see Supplementary Note.
Phylogenetic and principal component analyses of EBV genome sequences
The phylogenetic and principal component analyses were performed using EBV isolates sequenced by the current study and publicly accessible EBV genomes. For the phylogenetic analysis, we first created the fasta sequence for each resequenced isolate using the variant data extracted from the variant calling. The 230 EBV single-infection whole genomes were subsequently combined with the 97 public genomes and multiple sequence alignment was carried out using the multiple alignment program MAFFT42. After masking the regions of repetitive sequences and poor coverage in resequencing, the maximum likelihood of the phylogenetic relationship was inferred using the Randomized Axelerated Maximum Likelihood (RAxML) assuming a General Time Reversible (GTR) model43. The inferred phylogeny was subsequently rooted using the Evolutionary Placement Algorithm (EPA) algorithm44 from RAxML using a Macacine herpesvirus 4 genome sequence (NC_006146) as the outgroup.
In the PCA analysis, genomic variation from the 97 public genomes was generated by global pairwise sequence alignment of published genome sequences against the B95–8 reference genome (NC_007605.1) using the EMBOSS Stretcher45. The variant set is then combined with the variation data extracted from the WGS. A combined set of 12,182 SNPs from the 270 newly sequenced isolates and 97 published ones were then used for the PCA analyses. During the PCA analysis, SNPs were first filtered by allele frequency (minor genotype frequency > 0.05) and LD (pruning with a pairwise correlation R2 value > 0.6 within a 1000-bp sliding window). In total, 495 SNPs were included in the PCA analysis using the R package “SNPRelate”46.
Principal component analysis of cases and controls
To assess the human population structure of the 156 cases and 47 healthy controls used for the EBV GWAS discovery phase, the human DNAs of these samples were genotyped using the OmniZhongHua-8 Chip (Illumina). After sample filtering by a series of criteria, (i) the calling rate (above 95%), (ii) SNP filtering by minor allele frequency (above 5%), (iii) Hardy-Weinberg equilibrium (P > 1×10−6), and (iv) LD-based SNP pruning (R2 < 0.1 and not within the five high-LD regions5), PCA analysis was performed using the PLINK (Version 1.9) based on the discovery samples alone or by combining them with reference samples from the 1000 Genome project20.
Association analysis
Genetic associations of EBV variants were analyzed by testing either single or multiple variants. Single-variant association analysis used a generalized-linear mixed model with EBV genetic relatedness matrix as random effects21. Sex and age were included as fixed effects, as well as four human PCs and previously reported human NPC GWAS loci (rs2860580 and rs2894207) at HLA locus to correct for any potential impact of human population structures and genetics on the association results. Both single- and multiple-infection samples were included in the association analysis with the status of single- or multiple-infection being a covariate to correct for any potential confounding effect of multiple infections. The genome-wide discovery analysis was performed by testing 1,545 EBV variants (with missing rate < 10%, minor genotype frequency > 0.05, and heterozygosity < 0.1) in 156 cases and 47 healthy controls. The validation analysis was performed by testing three EBV non-synonymous coding SNPs 162215C>A, 162476T>C, and 163364C>T in BALF2 in an additional 483 cases and 605 population controls matched to the cases by age and sex from the case-control study in Zhaoqing County. The logistic regression model was used for validation, adjusting for age, sex, the human SNPs (rs2860580 and rs2894207 in HLA locus) and the status of single- or multiple-infection of EBV. The meta-analysis of the discovery and validation phases was performed with the z-score pooling method. Considering the extensive LD across the EBV genome, to obtain a suggestive genome-wide significance of association, we used permutations of a logistic model adjusting for age, sex, status of single- or multiple-infection, and the human and EBV population structures. The genome-wide significance (4.07×10−4) was determined with a 5% quantile of the empirical distribution of minimum P-values from 10, 000 permutations as the data-driven threshold to control family-wise error rate under multiple correlated testing.
The genome-wide multi-variant-based association analysis was performed by testing 1477 bi-allelic EBV variants in Bayesian variable selection regression implemented in piMASS22. Age, sex, four human PCs, two EBV PCs and the human SNPs (rs2860580 and rs2894207) were included as covariates. The analysis was performed by partitioning the EBV genome into the regions of a 20-SNP sliding window with 10 overlapping SNPs. The sum of the posterior probabilities of the SNPs being associated within a window was calculated as the “region statistic” indicating the strength of the evidence for genetic associations in that region.
To further prioritize potentially casual SNPs in the top hit BALF2 gene region for validation, we applied further fine-mapping analysis using Bayesian multiple-variable selection by PAINTOR3.147. Functional annotation of SNPs was used as the prior to compute the probability of being causal for each variant in the region. We assumed a single causal variant in BALF2 genes and calculated a 95% credible set that contains the minimum set of variants jointly having at least a 95% probability of including the causal variant.
We also evaluated the association of seven previously reported human GWAS SNPs with NPC in our combined samples of 639 cases and 652 controls. Of the seven SNPs, two within HLA locus, rs2860580 and rs2894207, showed significant associations with consistent ORs after multiple testing correction (Supplementary Table 10). For the rest SNPs in HLA (rs28421666), CDKN2A/2B (rs1412829), TERT (rs31489), TNFRSF19 (rs9510787), and MECOM (rs6774494) loci, the ORs in our samples were consistent with the previously reported values, although their evidences did not reach statistical significance after multiple testing correction (Supplementary Table 10). Therefore, we have done the association analyses of EBV variants including the two significant human GWAS SNPs (rs2860580 and rs2894207) as covariates. The results with the two human SNPs are very similar to the results without them as covariates (Supplementary Table 20). These findings clearly indicate that the reported human GWAS loci do not affect our association evidences for the EBV risk variants. A Life Sciences Reporting Summary for this paper is available.
Estimation of the population attributable fraction of risk
The proportion of NPC risk explained by the effect of the two high-risk haplotypes of SNPs 162476T>C and 163364C>T (C-T and C-C) was estimated in the validation sample. The attributable fraction of risk and 95% confidence interval were estimated in a logistic regression model adjusting for age and sex with the R package ‘AF’48. Because NPC is not a common disease (prevalence < 40/100,000), the risk ratio can be approximated by OR. Thus, the population attributable fraction can be approximated by
Data availability
The EBV sequencing data are deposited in NCBI database under BioProject ID PRJNA522388. EBV sequences are released in NCBI database under GenBank ID MK540241-MK540470.
Supplementary Material
Acknowledgements
We thank all the participants for their generous support for the current study. We would also thank R. Sun, C. Wang, H. Chen, J. Shen and C. Jie for helpful discussions on viral biology and genetic statistical, evolution and phylogenetic analyses, W.-S. Liu and X. Zuo for providing code supports, Z. Lin from Tulane University for kindly sharing EBV genome annotation files, and J.-Y. Shao from Sun Yat-sen University Cancer Center for providing MassArray iPlex platform.
This work was supported by the National Natural Science Foundation of China (81430059 to Y.-X.Z. and 81872228 to M.X.), the National Key R&D Program of China (No. 2016YF0902000 to Y.-X.Z.; 2018YFC1406902 and 2018YFC0910400 to W.Z.), the National Cancer Institute at the US NIH and NIH (R01CA115873-01 to H.-O.A. and Y.-X.Z.; R35-CA197449, P01-CA134294, U01-HG009088, U19-CA203654 to X.L.), and the Agency of Science, Technology and Research (A*STAR), Singapore (to J.L.).
Footnotes
Competing interests
The authors have no conflicts of interest to disclose.
References
- 1.Epstein MA, Achong BG & Barr YM Virus Particles in Cultured Lymphoblasts from Burkitt’s Lymphoma. Lancet 1, 702–3 (1964). [DOI] [PubMed] [Google Scholar]
- 2.Epstein A Why and How Epstein-Barr Virus Was Discovered 50 Years Ago. Curr Top Microbiol Immunol 390, 3–15 (2015). [DOI] [PubMed] [Google Scholar]
- 3.Kieff ED & Rickinson AB Epstein-Barr Virus and Its Replication. in Fields’ virology Vol. 68A (eds. Knipe DM & Howley PM) 2603–2654 (Lippincott Williams & Wilkins, Wolters Kluwer, Philadelphia, 2007). [Google Scholar]
- 4.Zhang LF et al. Incidence trend of nasopharyngeal carcinoma from 1987 to 2011 in Sihui County, Guangdong Province, South China: an age-period-cohort analysis. Chin J Cancer 34, 350–7 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bei JX et al. A genome-wide association study of nasopharyngeal carcinoma identifies three new susceptibility loci. Nat Genet 42, 599–603 (2010). [DOI] [PubMed] [Google Scholar]
- 6.Bei JX et al. A GWAS Meta-analysis and Replication Study Identifies a Novel Locus within CLPTM1L/TERT Associated with Nasopharyngeal Carcinoma in Individuals of Chinese Ancestry. Cancer Epidemiol Biomarkers Prev 25, 188–192 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cui Q et al. An extended genome-wide association study identifies novel susceptibility loci for nasopharyngeal carcinoma. Hum Mol Genet (2016). [DOI] [PubMed] [Google Scholar]
- 8.Tang M et al. The principal genetic determinants for nasopharyngeal carcinoma in China involve the HLA class I antigen recognition groove. PLoS Genet 8, e1003103 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Baer R et al. DNA sequence and expression of the B95–8 Epstein-Barr virus genome. Nature 310, 207–11 (1984). [DOI] [PubMed] [Google Scholar]
- 10.Zeng MS et al. Genomic sequence analysis of Epstein-Barr virus strain GD1 from a nasopharyngeal carcinoma patient. J Virol 79, 15323–30 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dolan A, Addison C, Gatherer D, Davison AJ & McGeoch DJ The genome of Epstein-Barr virus type 2 strain AG876. Virology 350, 164–70 (2006). [DOI] [PubMed] [Google Scholar]
- 12.Liu P et al. Direct sequencing and characterization of a clinical isolate of Epstein-Barr virus from nasopharyngeal carcinoma tissue by using next-generation sequencing technology. J Virol 85, 11291–9 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lin Z et al. Whole-genome sequencing of the Akata and Mutu Epstein-Barr virus strains. J Virol 87, 1172–82 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Palser AL et al. Genome diversity of epstein-barr virus from multiple tumor types and normal infection. J Virol 89, 5222–37 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Correia S et al. Natural Variation of Epstein-Barr Virus Genes, Proteins, and Primary MicroRNA. J Virol 91(2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kwok H et al. Genomic diversity of Epstein-Barr virus genomes isolated from primary nasopharyngeal carcinoma biopsy samples. J Virol 88, 10662–72 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Edwards RH, Seillier-Moiseiwitsch F & Raab-Traub N Signature amino acid changes in latent membrane protein 1 distinguish Epstein-Barr virus strains. Virology 261, 79–95 (1999). [DOI] [PubMed] [Google Scholar]
- 18.Hui KF et al. High risk Epstein-Barr virus variants characterized by distinct polymorphisms in the EBER locus are strongly associated with nasopharyngeal carcinoma. Int J Cancer (2018). [DOI] [PubMed] [Google Scholar]
- 19.Tso KK et al. Complete genomic sequence of Epstein-Barr virus in nasopharyngeal carcinoma cell line C666–1. Infect Agent Cancer 8, 29 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Genomes Project C et al. A global reference for human genetic variation. Nature 526, 68–74 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen H et al. Control for Population Structure and Relatedness for Binary Traits in Genetic Association Studies via Logistic Mixed Models. Am J Hum Genet 98, 653–66 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Guan Y & Stephens M Bayesian variable selection regression for genome-wide association studies and other large-scale problems. Ann. Appl. Stat 5, 1780–1815 (2011). [Google Scholar]
- 23.Decaussin G, Leclerc V & Ooka T The lytic cycle of Epstein-Barr virus in the nonproducer Raji line can be rescued by the expression of a 135-kilodalton protein encoded by the BALF2 open reading frame. J Virol 69, 7309–14 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zeng Y, Middeldorp J, Madjar JJ & Ooka T A major DNA binding protein encoded by BALF2 open reading frame of Epstein-Barr virus (EBV) forms a complex with other EBV DNA-binding proteins: DNAase, EA-D, and DNA polymerase. Virology 239, 285–95 (1997). [DOI] [PubMed] [Google Scholar]
- 25.Mumtsidu E et al. Structural features of the single-stranded DNA-binding protein of Epstein-Barr virus. J Struct Biol 161, 172–87 (2008). [DOI] [PubMed] [Google Scholar]
- 26.Rowe M et al. Distinction between Epstein-Barr virus type A (EBNA 2A) and type B (EBNA 2B) isolates extends to the EBNA 3 family of nuclear proteins. J Virol 63, 1031–9 (1989). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li DJ et al. The dominance of China 1 in the spectrum of Epstein-Barr virus strains from Cantonese patients with nasopharyngeal carcinoma. J Med Virol 81, 1253–60 (2009). [DOI] [PubMed] [Google Scholar]
- 28.Coghill AE et al. Identification of a Novel, EBV-Based Antibody Risk Stratification Signature for Early Detection of Nasopharyngeal Carcinoma in Taiwan. Clin Cancer Res 24, 1305–1314 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Paramita DK et al. Native early antigen of Epstein-Barr virus, a promising antigen for diagnosis of nasopharyngeal carcinoma. J Med Virol 79, 1710–21 (2007). [DOI] [PubMed] [Google Scholar]
- 30.Steven NM et al. Immediate early and early lytic cycle proteins are frequent targets of the Epstein-Barr virus-induced cytotoxic T cell response. J Exp Med 185, 1605–17 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xue WQ et al. Decreased oral Epstein-Barr virus DNA loads in patients with nasopharyngeal carcinoma in Southern China: A case-control and a family-based study. Cancer Med (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hadinoto V, Shapiro M, Sun CC & Thorley-Lawson DA The dynamics of EBV shedding implicate a central role for epithelial cells in amplifying viral output. PLoS Pathog 5, e1000496 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
Methods-only references
- 33.Li H & Durbin R Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25, 1754–60 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li H et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25, 2078–9 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.DePristo MA et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet 43, 491–8 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cingolani P et al. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly (Austin) 6, 80–92 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Raab-Traub N & Flynn K The structure of the termini of the Epstein-Barr virus as a marker of clonal cellular proliferation. Cell 47, 883–9 (1986). [DOI] [PubMed] [Google Scholar]
- 38.Pathmanathan R, Prasad U, Sadler R, Flynn K & Raab-Traub N Clonal proliferations of cells infected with Epstein-Barr virus in preinvasive lesions related to nasopharyngeal carcinoma. N Engl J Med 333, 693–8 (1995). [DOI] [PubMed] [Google Scholar]
- 39.Neri A et al. Epstein-Barr virus infection precedes clonal expansion in Burkitt’s and acquired immunodeficiency syndrome-associated lymphoma. Blood 77, 1092–5 (1991). [PubMed] [Google Scholar]
- 40.Weiss ER et al. Early Epstein-Barr Virus Genomic Diversity and Convergence toward the B95.8 Genome in Primary Infection. J Virol 92(2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Browning SR & Browning BL Rapid and accurate haplotype phasing and missing-data inference for whole-genome association studies by use of localized haplotype clustering. Am J Hum Genet 81, 1084–97 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Katoh K, Misawa K, Kuma K & Miyata T MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res 30, 3059–66 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stamatakis A RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22, 2688–90 (2006). [DOI] [PubMed] [Google Scholar]
- 44.Berger SA, Krompass D & Stamatakis A Performance, accuracy, and Web server for evolutionary placement of short sequence reads under maximum likelihood. Syst Biol 60, 291–302 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li W et al. The EMBL-EBI bioinformatics web and programmatic tools framework. Nucleic Acids Res 43, W580–4 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zheng X et al. A high-performance computing toolset for relatedness and principal component analysis of SNP data. Bioinformatics 28, 3326–8 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kichaev G et al. Integrating functional data to prioritize causal variants in statistical fine-mapping studies. PLoS Genet 10, e1004722 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dahlqwist E, Zetterqvist J, Pawitan Y & Sjolander A Model-based estimation of the attributable fraction for cross-sectional, case-control and cohort studies using the R package AF. Eur J Epidemiol 31, 575–82 (2016). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The EBV sequencing data are deposited in NCBI database under BioProject ID PRJNA522388. EBV sequences are released in NCBI database under GenBank ID MK540241-MK540470.