Figure 4. Vκ rearrangement during B cell development and repertoire analysis of E88∆ mice.
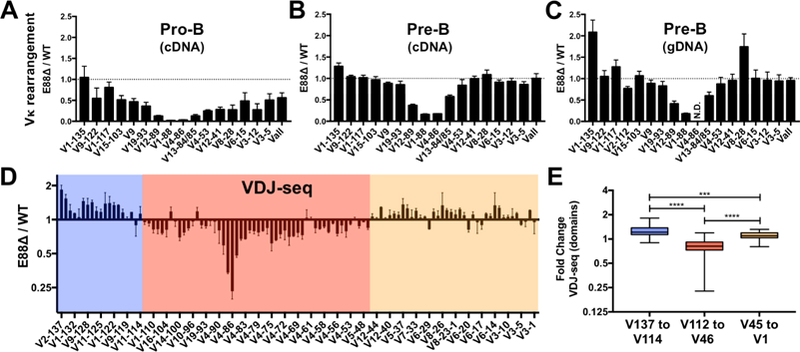
(A-C) BM-derived CD19+ cells from WT and E88∆ mice were purified and sorted for pro-B cells and small pre-B cells. Pro-B cell RNA (A), small pre-B cell RNA (B) and small pre-B cell gDNA (C) were harvested and analyzed for Vκ rearrangement by qPCR. Data was normalized to GAPDH for RNA and to a DNA-loading control (Eµ) for gDNA and is shown as a ratio of E88∆ / WT. N.D.= not detected for WT or E88∆. (D) Small pre-B cell gDNA was analyzed by deep sequencing (VDJ-seq). Data is the average of two experiments and is expressed as the ratio of E88∆ / WT of the percentage of total reads per sample ± SEM. Domains where the majority of the Vκ gene rearrangements are underrepresented (pink box) or overrepresented (blue and tan boxes) in the E88∆ mice are indicated. (E) VDJ-seq results were separated into 3 groups corresponding to the 3 colored regions in panel D and analyzed. T-test was used to calculate significant differences between groups (***p<0.001, and ****p<0.0001).
