Figure 6. LRCI in the Igκ locus are altered by E88 deletion at the pro-B cell stage.
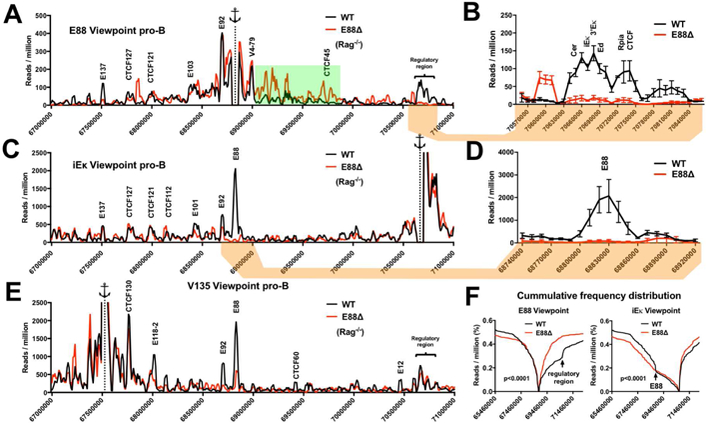
4C libraries from WT.Rag−/− and E88∆.Rag−/− CD19+ pro-B cells were assessed by deep sequencing. (A, C, E) 4C interactions plots for the Igκ locus from E88, iEκ, or E135 viewpoints for WT (black) or E88∆ (red). The interaction frequencies are expressed as reads per million (RPM) in 10 kb windows using a running window of 30 kb for the indicated genomic locations. The anchor represents the viewpoint. Green highlight indicates an area with higher density of specific LRCI in E88∆ pro-B cells. (B, D) Close-up view of indicated areas showing the average of the 3 experiments ± SEM. (F) Cumulative frequency distributions (CFD) from E88 and iEκ viewpoints show the cumulative 4C reads in E88∆ compared to WT pro-B cells (p<0.0001). Mann-Whitney test was used to calculate the statistically significant difference between CFD from WT and E88∆. Data is the average of 3 independent biological samples. Significant differential 4C interactions between the compared cell types are shown in Table S3.
