Figure 1. Stromal RNA Is Transferred to Breast Cancer Cells by Exosomes.
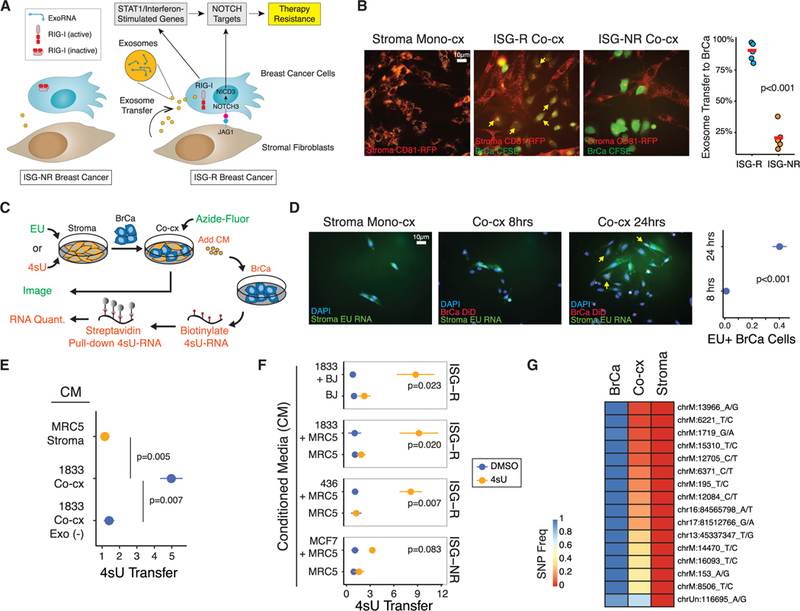
(A) Summary of ISG-R and ISG-NR breast cancer cells and differential exosome transfer and RIG-I activation upon interaction with stromal fibroblasts.
(B) MRC5 fibroblasts expressing a CD81-RFP exosome reporter were co-cultured with CFSE-labeled ISG-R 1833 or ISG-NR MCF7 breast cancer cells. Exosome transfer is quantitated (right) and representative transfer is shown (arrows).
(C) Schema for measuring RNA transfer from stromal to breast cancer cells utilizing the uridine analog EU for fluorescence microscopy (green) or 4sU for streptavidin pull-down (orange).
(D) MRC5 fibroblasts were labeled with EU and co-cultured with DiD lipid-labeled 1833 breast cancer cells. EU-positive 1833 cells (yellow arrows) and quantitation are shown.
(E) Relativetransferof4sU RNA to mono-cultured 1833 breast cancer cells after addition of conditioned media (CM) isolated from 4sU-labeled MRC5 fibroblasts grown in mono-culture (Stroma, orange) or from 1833 ISG-R co-culture (Co-cx, blue). Co-culture CM depleted of exosomes (Co-cx Exo(—) CM) is shown as a control for exosome-dependency (n = 5).
(F) Same as in (E) except CM was isolated from MRC5 or BJ 4sU-labeled fibroblasts grown in mono-culture or co-cultured with the indicated ISG-R or ISG-NR breast cancer cells. Shown is relative 4sU RNA transfer after CM addition to each breast cancer cell mono-culture(n = 3). Transfer is relative to mock4sU labeling using DMSO.
(G) Allelic frequency of exoRNA SNPs from exosomes isolated from 1833 breast cancer (BrCa), MRC5 fibroblasts (Stroma), or from co-culture of both cell types (Co-cx). Analysis is based on SNPs present in exoRNA from breast cancer cells and not present in fibroblasts. Error bars are SEM of biological replicates. See also Figure S1.
