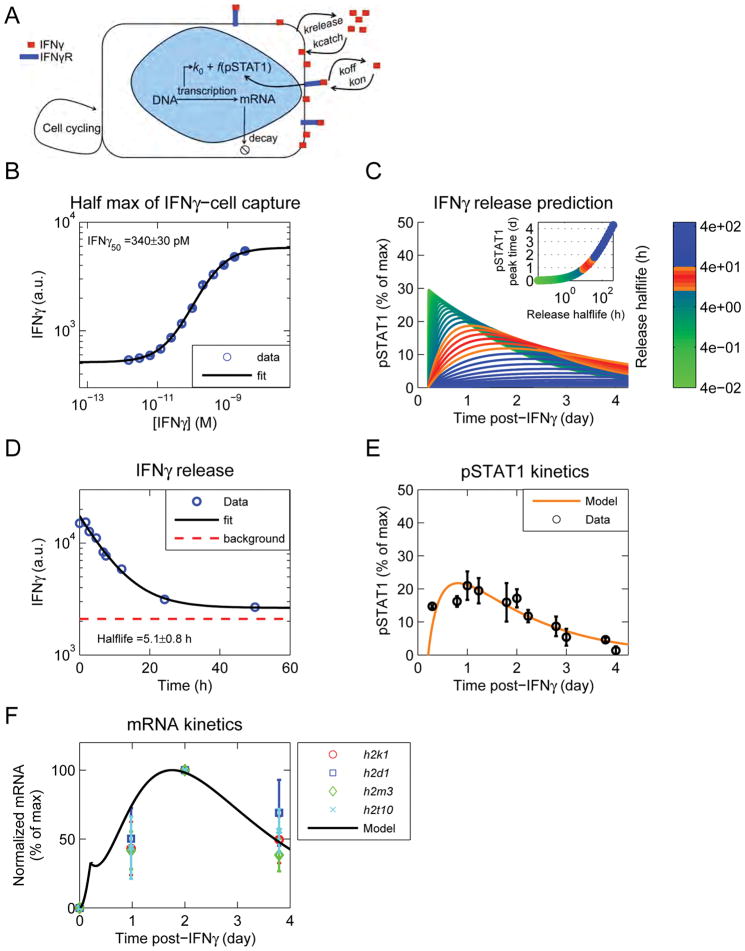
Figure 4. A mathematical model, including a slow catch and release process, recapitulates experimental results.
See also Figure S4. (A) Schematic of model. IFNγ binds to the IFNγR and is captured by the cells in a receptor-independent manner. After removal of exogenous IFNγ, release from cells drives persistent signaling through the IFNγR, until it is consumed. (B) B16 IFNγR KO cells were exposed to different doses of IFNγfluo, washed, and fluorescence was quantified by flow cytometry. The data were fit with a Hill function and the IFNγ50 was computed from the fit. Data are representative of 3 independent experiments. (C) The model was run keeping the IFNγ50 constant, and varying the catch and release rates. pSTAT1 was calculated from the EC50 of signaling (Figure S4A) and the concentration of free IFNγ generated from cell release (Figure S4D). Curves colored in orange and red resemble the pSTAT1 curves observed experimentally (Figure 2B). Inset shows the time of pSTAT1 peak versus the halflife of cell release. (D) B16 IFNγR KO cells were pulsed with IFNγfluo in a large volume of well mixed RPMI. The media was then replaced with an excess of unlabeled IFNγ and the decay of cell fluorescence was quantified by flow cytometry. The data were fit with an exponential decay function and the decay rate was computed from the fit. Data are representative of 3 independent experiments. (E) The model was updated with the IFNγ-cell release rate. The cell catch rate was inferred from: (IFNγ50=krelease/kcatch). The model was fit to the pSTAT1 curve obtained after removal of exogenous IFNγ (Figure 2B). (F) The pSTAT1 curve obtained from the model in figure 5E was used to calculate the mRNA trajectory and compared to several candidate genes from Figure 1E, cluster 1.