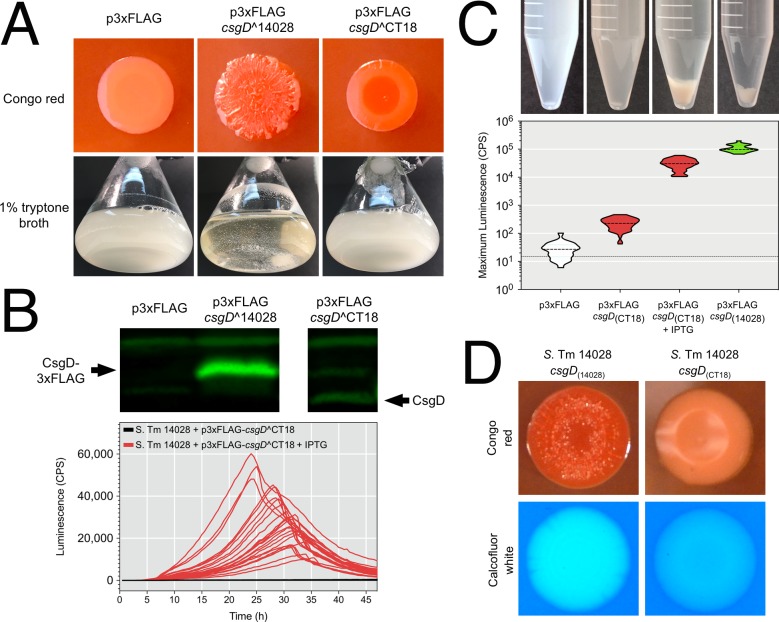
Fig 7. Functional analysis of the S. Typhi CT18 csgD allele.
(A) The csgD alleles from S. Typhimurium 14028 and S. Typhi CT18 were cloned into p3xFLAG and transformed into S. Typhimurium 14028 ΔcsgD cells. Colony morphology and flask cultures of uninduced cells were evaluated for biofilm phenotypes. (B) Top panel: Whole cell lysates were generated from cells acquired from flask cultures and probed for synthesis of CsgD via Western blot. Bottom panel: A csgB promoter-reporter construct was used to evaluate CsgD activity in S. Typhimurium 14028 ΔcsgD cells harbouring p3xFLAG-csgD^CT18 with or without IPTG induction. (C) Top panel: Presence or absence of multicellular aggregates in flask cultures of S. Typhimurium 14028 ΔcsgD cells containing p3xFLAG constructs (+/- IPTG) or p3xFLAG alone. Bottom panel: Maximum promoter activity of a csgB promoter-reporter construct measured in microbroth cultures corresponding to the tubes shown in the top panel. (D) Biofilm phenotypes of S. Typhimurium 14028 strains that contained the native csgD^14028 allele or the truncated csgD allele from S. Typhi CT18 following genome engineering.