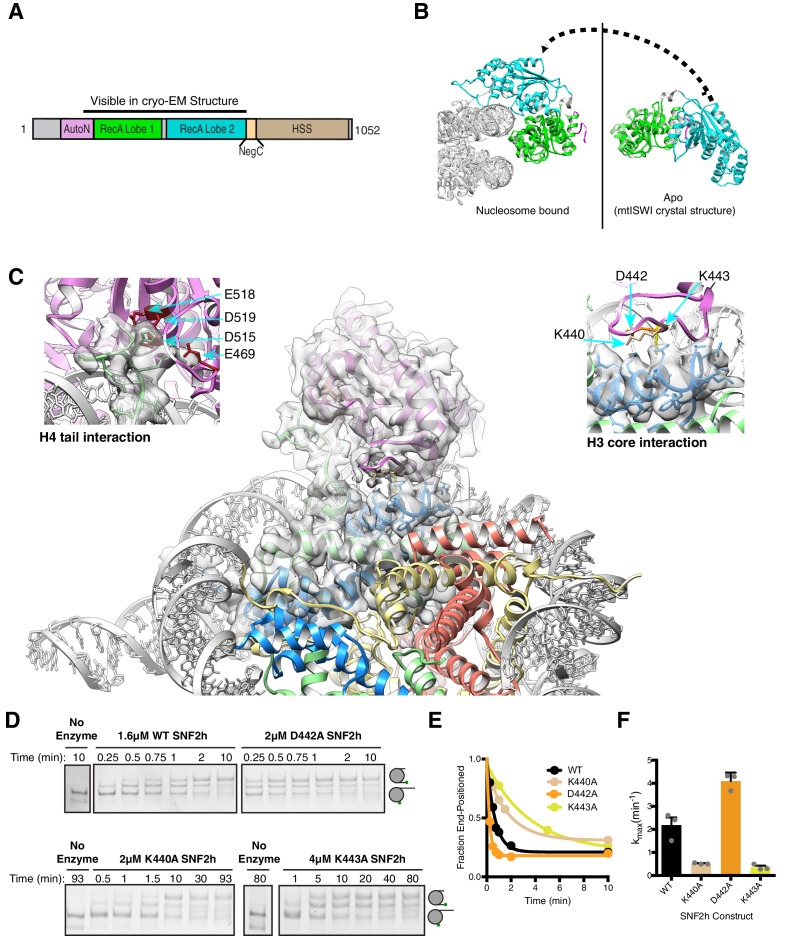
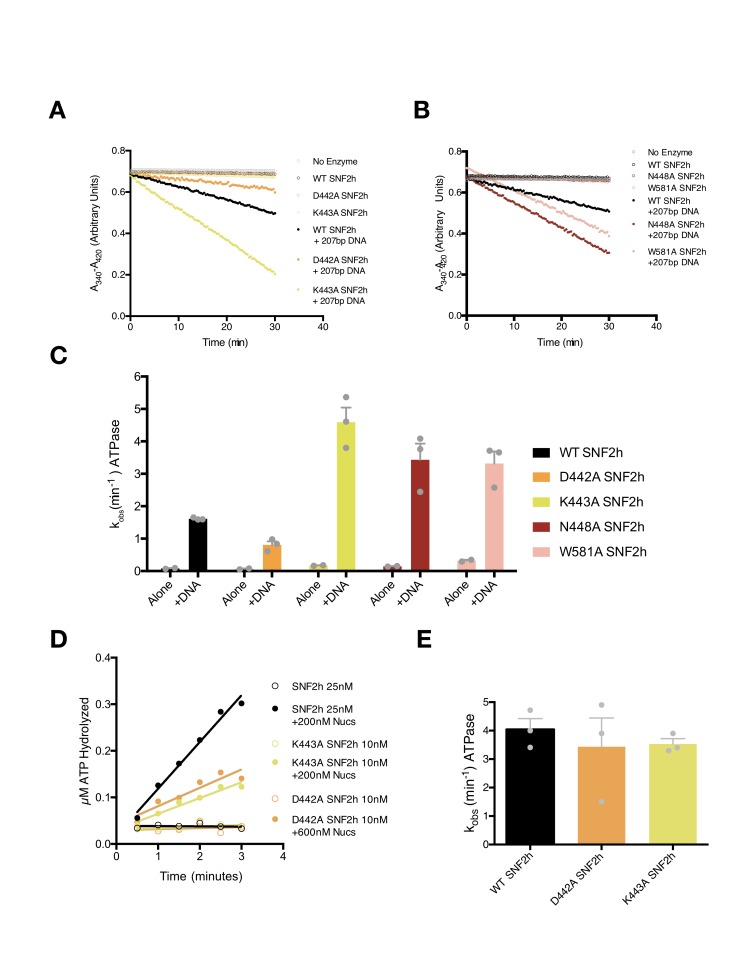
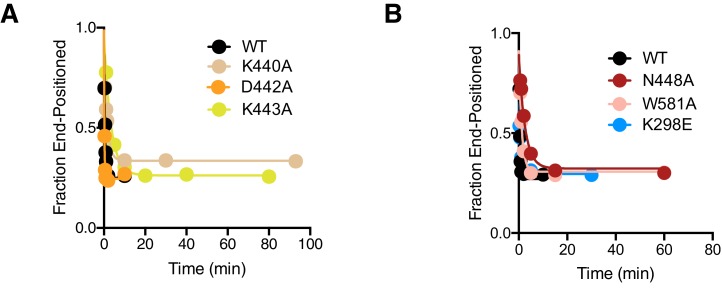
Figure 3. Interactions of SNF2h with the histone proteins.
(A) Domain diagram of SNF2h. (B) Conformational changes in SNF2h associated with nucleosome binding. SNF2h is colored according to the domain diagram. The apo structure is the Myceliophtora thermophila ISWI crystal structure (PDBID: 5JXR). (C) Middle. High resolution SNF2h-nucleosome structure from Figure 1 enlarged to show details of the interactions with the histone proteins. Colored in red on SNF2h are the acidic residues contacting the histone H4 tail. Colored in orange, tan, and yellow are the residues mutated in this study. Left. Enlarged to show details of the H4 tail interaction. Right. Enlarged to show details of the H3 core interaction. (D) Native gel remodeling assay of SNF2h constructs. Cy3-DNA labeled nucleosomes were incubated with saturating concentrations of enzyme and ATP and resolved on a native 6% polyacrylamide gel. (E) Quantifications of the data in (D) zoomed on the x-axis to show effects more clearly and fit to a single exponential decay. Un-zoomed plots are in Figure 3—figure supplement 6. (F) Rate constants derived from remodeling assays. Bars represent the mean and standard error from three experiments.