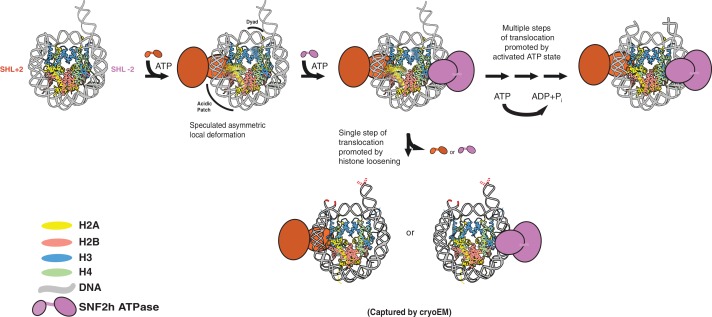
Figure 5. Speculative model that places SNF2h-nucleosome Cryo-EM structures within SNF2h reaction cycle .
Two protomers of SNF2h bind to the nucleosome along with ATP. Based on previous work, the directionality of nucleosome sliding is determined by the motor that engages the longer flanking DNA (Leonard and Narlikar, 2015). By this model, the SNF2h motor bound at SHL+2 (orange protomer) will be the active motor and determine the direction of sliding because it would contact the 60 bp flanking DNA with its HSS domain (Leonard and Narlikar, 2015). For simplicity, the HSS domain is not shown. Binding of the SHL+2 protomer asymmetrically deforms the acidic patch and histone H3 near the dyad on the opposite face of the histone octamer. The second protomer can bind at SHL-2, but cannot act because deformation of the acidic patch inhibits its ability to slide nucleosomes. The SNF2h complex with the deformed octamer represents an intermediate that is poised for translocation. Processive DNA translocation is enabled by successive ATP hydrolysis cycles from this activated intermediate, moving DNA in 1–2 bp fundamental increments. We speculate that the cryo-EM structure captured at low salt represents the deformed intermediate, while the structure captured at high salt represents a collapsed product state in which the nucleosome is translocated by 2 bp (translocated bases are highlighted in red).