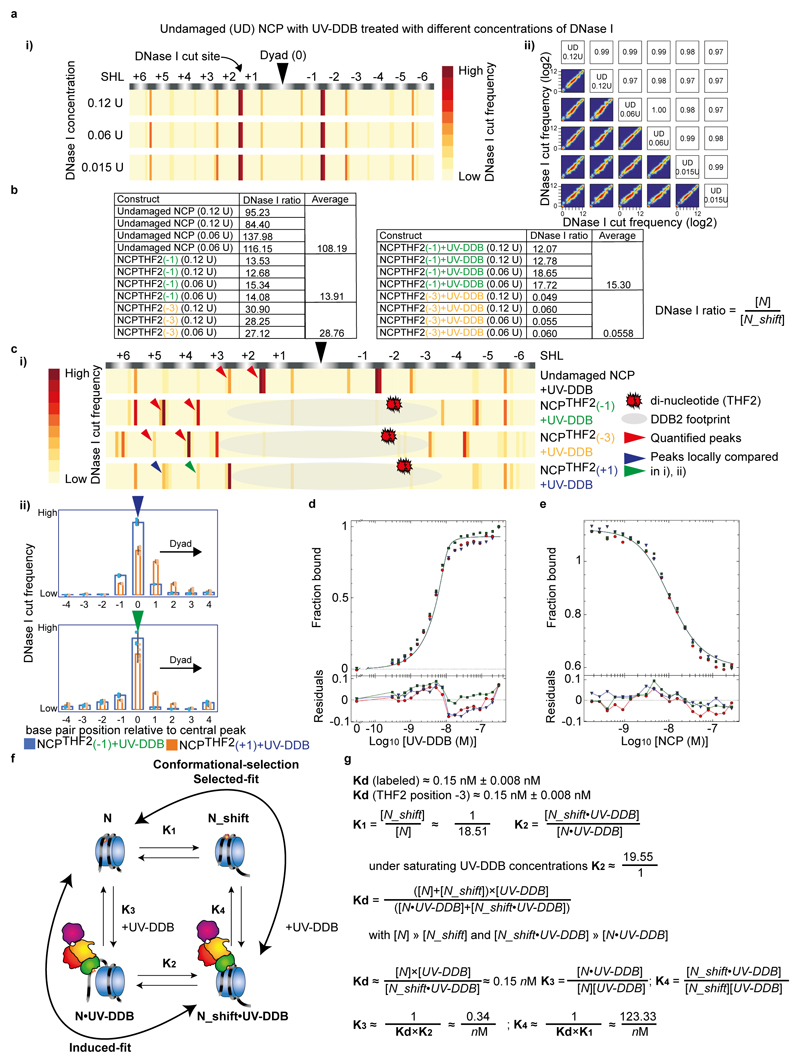
Extended Data Fig. 6. Thermodynamic dissection of UV-DDB binding with slide-assisted site-exposure (SAsSE).
a, DNase I digestion of undamaged (UD) nucleosomes with a range of enzyme concentrations (0.12 U to 0.015 U) show identical sensitive sites (i, data shown is the average of two replicates (n = 2) per enzyme concentration) and are highly reproducible across replicates (ii). Note that an example of the data reproducibility is shown for the UD construct, but the data for all constructs were highly reproducible (Person correlations of R = >0.95). b, Ratio of counts at position n versus n-3 for undamaged NCP, NCPTHF2(-1), NCPTHF2(-3) in the absence and presence of UV-DDB. c, i) Peaks for UD NCP, NCPTHF2(-1), NCPTHF2(-3) and NCPTHF2(+1) in the presence of UV-DDB used to quantify the register shift ratios (n=2). Note that in the case of NCPTHF2(+1) we expect only 30-40% of molecules to shift by maximal 1 bp. Our data are consistent with an increased population of α-satellite DNA in the +1 nucleosome shifted by 1 bp towards the dyad axis compared to the -1 nucleosome (ii, n = 4, mean ± s.d.), however, the width and the overlap of the shifted/unshifted peak prevented further detailed analysis. d, titration 10 nM of a Cy5-15-bp-DNATHF with increasing concentrations of UV-DDB. The resulting curve was fitted with Dynafit resulting in a Kd~ 0.15 nM. Given the tight Kd relative to the high concentration of the Cy5-15-bp-DNATHF label, the value should be viewed as estimate. These are in line, however, with previous experiments34. All data include three technical replicates (n = 3) and are shown as mean ± s.d.. e, Complex of 10 nM Cy5-15-bp-DNATHF and UV-DDB, was back titrated with NCPTHF2 (-3) and the data fitted numerically in Dynafit. All data include three technical replicates (n = 3) and are shown as mean ± s.d.. f, Thermodynamic binding scheme invoking induced fit binding, in which the register shift is induced on UV-DDB binding, and a conformational preselection branch (selected-fit), in which a preequilibrium exists that is pulled by UV-DDB binding. g, Equations describing the thermodynamic binding process, and approximations used to derive K1, K2, K3 and K4.