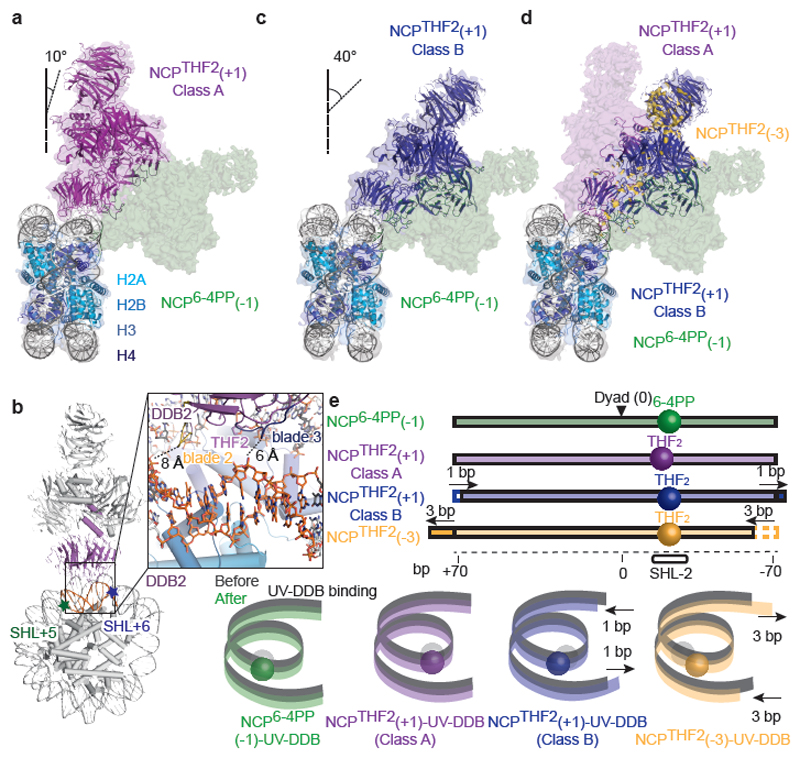
Figure 4. Subpopulations of translational settings revealed by UV-DDB binding.
a, Structure of NCPTHF2(+1)-UV-DDB Class A (magenta) (Extended Data Fig. 7) superimposed on NCP6-4PP(-1)-UV-DDB (green). b, Close-up of NCPTHF2(+1)-UV-DDB Class A showing the proximity of DDB2 to DNA between SHL5 and SHL6 (orange). c, Comparison of NCPTHF2(+1)-UV-DDB Class B (blue) and NCP6-4PP(-1)-UV-DDB (green). d, Superimposition of NCPTHF2(+1)-UV-DDB (Class A and Class B), NCPTHF2(-3)-UV-DDB (yellow), and NCP6-4PP(-1)-UV-DDB, showing bidirectional register shifting towards a common locus. e, Schematic representation of DNA with lesions indicated by spheres (sequences in Extended Data Fig. 8a).