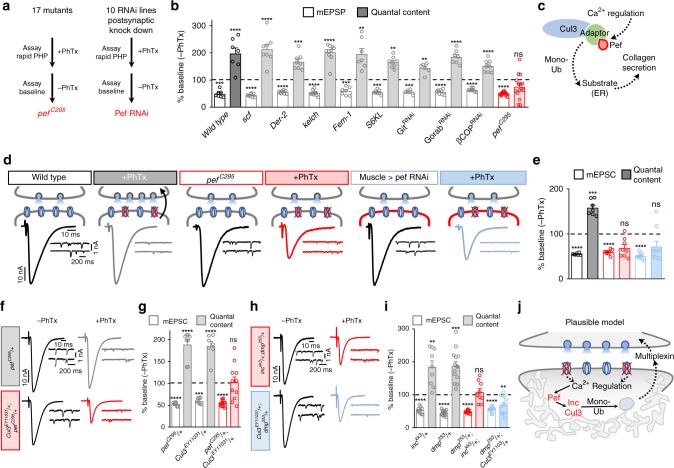
Fig. 6.
An Inc- and Cul3-interaction screen identifies peflin to be required postsynaptically for PHP expression. a Flow diagram and screening strategy of Inc- and Cul3-interacting genes identifies a mutation and RNAi line targeting Drosophila peflin (CG17765; pef). b Quantification of mEPSP and quantal content values in a subset of mutants screened normalized to baseline values (–PhTx) (n = 6–20; see Supplementary Table 1). c Schematic summarizing the role of mammalian Pef functioning as a co-adaptor for the Cul3-KLHL12 complex, which is regulated by Ca2+ signaling. This signaling activates Cul3KLHL12 to mono-ubiquitinate substrates involved in membrane trafficking at the ER to modulate Collagen secretion. d Schematic and representative traces of wild type, peflin mutants (pefC295), and Pef RNAi driven in the postsynaptic muscle (muscle > Pef RNAi: G14-Gal4/UAS-Pef RNAi) before and following PhTx application. Note that while mEPSC amplitude is reduced in all three genotypes after PhTx application, PHP fails to be expressed in pefC295 and muscle > Pef RNAi. e Quantification of mEPSC and quantal content values in the indicated genotypes normalized to baseline values (–PhTx: wild type, n = 8; pefC295, n = 7; muscle > Pef RNAi, n = 10; + PhTx: wild type, n = 7; pefC295, n = 8; muscle > Pef RNAi, n = 9). Representative traces (f and h) and quantification (g and i) of genetic interaction experiments between Cul3EY11031/pefC295 (Cul3EY11031/ + , n = 8 ( – PhTx, + PhTx); pefC295/ + , n = 8; Cul3EY11031/ + ,pefC295/ + , n = 21), inckk3/dmp253 (inckk3/ + , n = 8; dmp253/ + , n = 8; inckk3/ + ;dmp253/ + , n = 9) and Cul3EY11031/dmp253 (Cul3EY11031/ + ;dmp253/ + , n = 10). j Schematic illustrating a speculative but plausible model linking Cul3/Inc and Pef in retrograde homeostatic signaling. Asterisks indicate statistical significance using a Student’s t-test: (**) p < 0.01; (***) p < 0.001; (****) p < 0.0001, (ns) not significant. Error bars indicate ± SEM. n values indicate biologically independent cells. Additional statistical information and absolute values for normalized data can be found in Supplementary Table 2. Source data are provided as a Source Data file