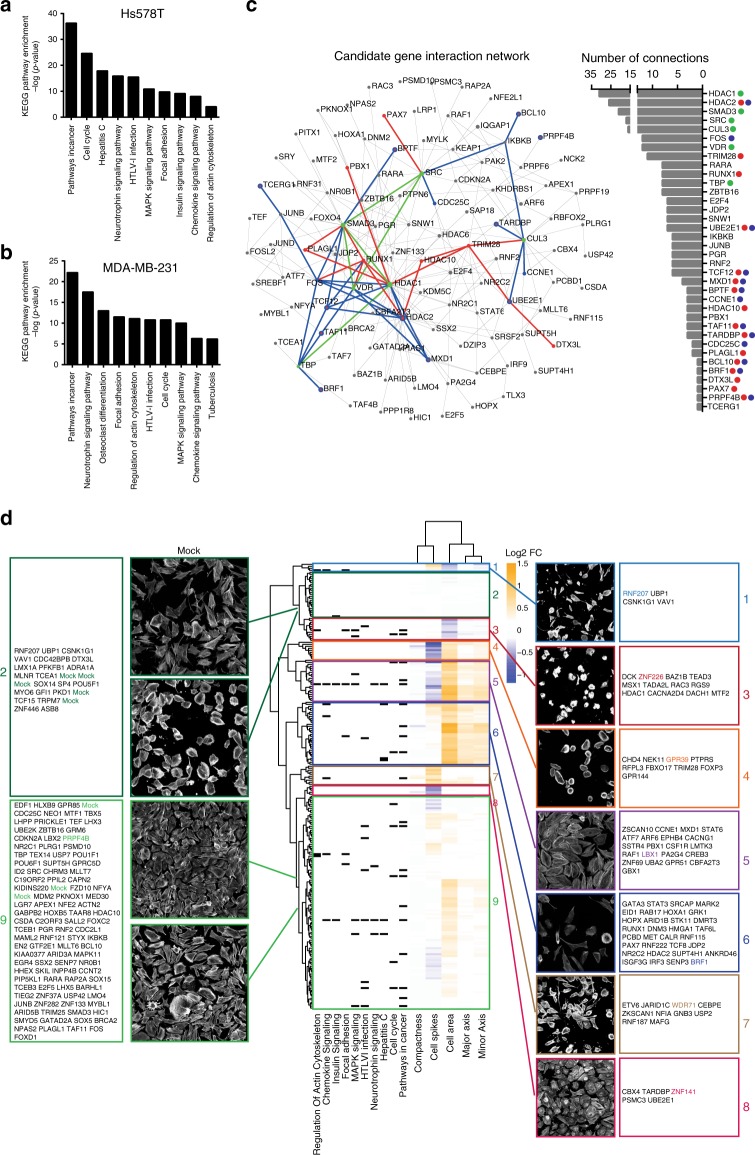
Fig. 4.
Regulatory networks drive tumor cell migration. a Enrichment of KEGG pathways in PPI networks generated from Hs578T candidate genes and b MDA-MB-231 candidate genes. NetworkAnalyst was used to generate PPI networks. c Zero-order interaction network of combined Hs578T and MDA-MB-231 candidate genes reveals a highly connected sub-network of clinically associated genes (in blue). Candidate genes inhibiting cell migration in both cell lines are shown in red; central hubs are highlighted in green. The degree of connectivity (number of connections) is displayed on the right. d Phenotype-based clustering of the PKT validated candidate genes based on morphological changes in the Hs578T cell line. Per parameter, log2 fold change (FC) compared to mock control was calculated. Clustering was performed based on Euclidean distance and complete linkage