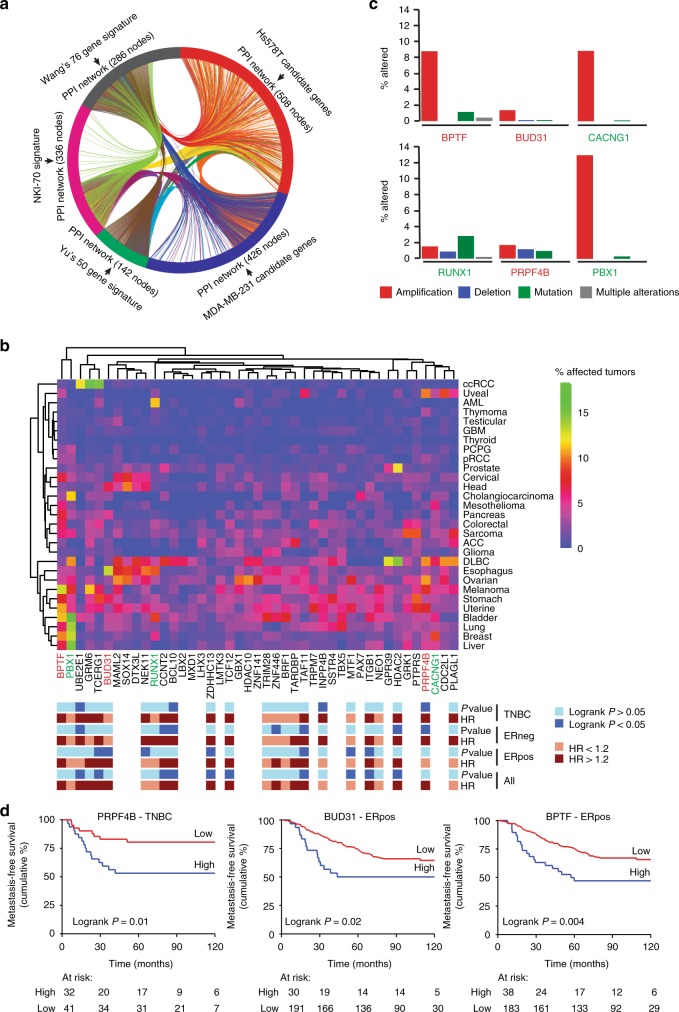
Fig. 5.
Modulators of TNBC cell migration are related to BC metastasis-free survival. a Prognostic gene signatures were used to generate minimum interaction PPI networks and compared to our candidate TNBC cell migration gene networks. Candidate genes affecting cell migration feed into similar networks essential for BC progression and metastasis formation. b Hierarchical clustering (Euclidean distance, complete linkage) of genetic modifications (mutations, deletions and amplifications combined) of 43 candidate genes in 29 cancer types. Data was derived from The Cancer Genome Atlas. Annotation shows the expression of the candidates in relation to BC metastasis-free survival in different BC subtypes. P-values were calculated using Cox proportional hazards regression analysis, with gene expression values as continuous variable and metastasis-free survival as end point. Genes marked in red and blue are highlighted in c. Red genes were selected for further analysis. c Contribution of different genetic modifications to the rate in several highly mutated or amplified candidates. d Kaplan–Meier curves of for expression of PRPF4B, BUD31, and BPTF and relation to metastasis-free survival in ERpos or TNBC breast cancer. Gene expression data of lymph-node-negative BC patient cohort without prior treatment using optimal split was used to obtain the curves