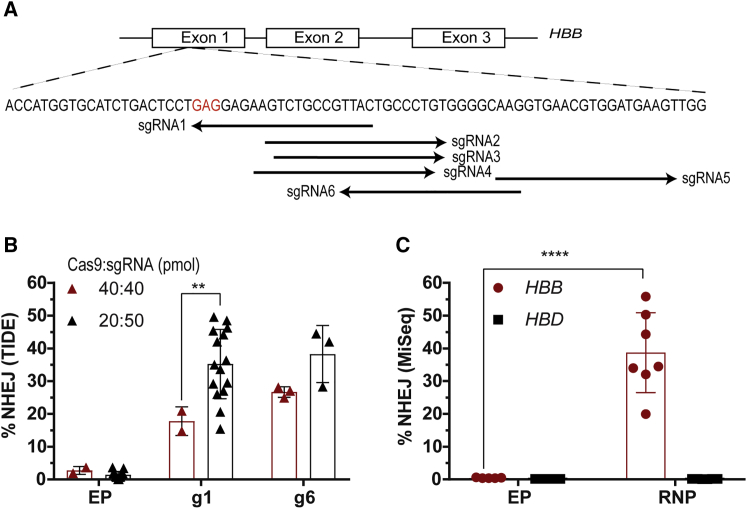
Figure 1.
Screening of Nucleases to Create DSBs in Exon 1 of the HBB Gene
(A) Schematic representation of the genomic HBB gene showing the location of sgRNA-binding sites. A nucleotide substitution from GAG (codon 6 in red) to GTC or GTG changes the amino acid from glutamate to valine and causes SCD. (B) Optimizing the Cas9:sgRNA ratio to maximize editing efficiency in mPBSCs. NHEJ rates were analyzed by TIDE/ICE sequencing (Cas9:sgRNA ratio of 1:1 [40 pmol each], donor n = 2 or ratio of 1:2.5 [20 pmol of Cas9 and 50 pmol of sgRNA], donor n = 15). (C) Evaluating on-target disruption at HBB and possible off-target disruption at HBD by MiSeq analysis in mPBSCs using sgRNA-g1 delivered as RNP (donor n = 7). All bar graphs show mean ± SD. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001. p value was calculated by comparing each sample mean with the respective control sample mean by two-way ANOVA with Dunnett’s multiple comparison. See also Figure S1 and Table S2.