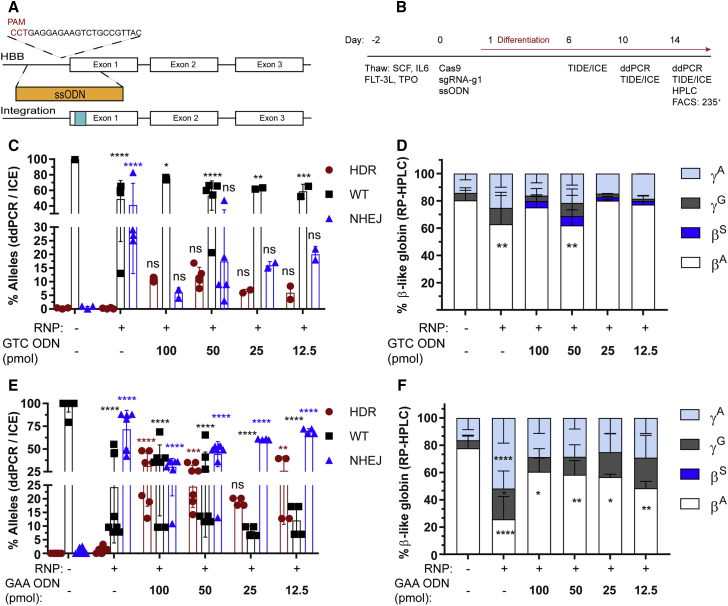
Figure 3.
Homology-Directed Repair at the HBB Nuclease Target Site Using ssODN Donor Template
(A) Schematic representation of ssODN cassette designed to drive either a GTC (E6V) introducing a sickle mutation or a GAA (E6optE) introducing a codon-optimized change at codon 6 by HDR. (B) Experimental timeline for testing gene editing with RNP and ssODN delivery followed by erythroid differentiation in mPBSCs. (C) WT (%) and HDR (%) measured by ddPCR and NHEJ (%) measured by ICE sequencing, respectively, following electroporation with RNP alone or co-delivery of RNP and GTC (E6V) ssODN donor template, at the indicated concentrations (50 pmol ssODNs, donor n = 5). (D) RP-HPLC analysis of erythroid cells to measure β-globin expression in cells treated with RNP only or RNP plus GTC (E6V) ssODNs (50 pmol ssODNs, donor n = 5). (E) WT (%) and HDR (%) measured by ddPCR and NHEJ (%) measured by ICE sequencing, respectively, following electroporation with RNP alone or co-delivery of RNP and GAA (E6optE) ssODNs, at the indicated concentrations (50 pmol ssODNs, donor n = 8). (F) RP-HPLC analysis of erythroid cells to measure β-globin expression in cells treated with RNP only or RNP plus GAA (E6optE) ssODNs (50 pmol ssODNs, donor n = 6). α, alpha; βA, adult; βS, sickle; γG, gamma 2; γA, gamma 1. All bar graphs show mean ± SD. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001. p value was calculated by comparing each sample mean of NHEJ (%), HDR (%), WT (%), or globin sub-type (%) with the respective NHEJ (%), HDR (%), WT (%), or % globin sub-type (%) of the mock sample by two-way ANOVA with Dunnett’s multiple comparison. Asterisks are color matched to the respective mock sample. See also Figures S5–S8.