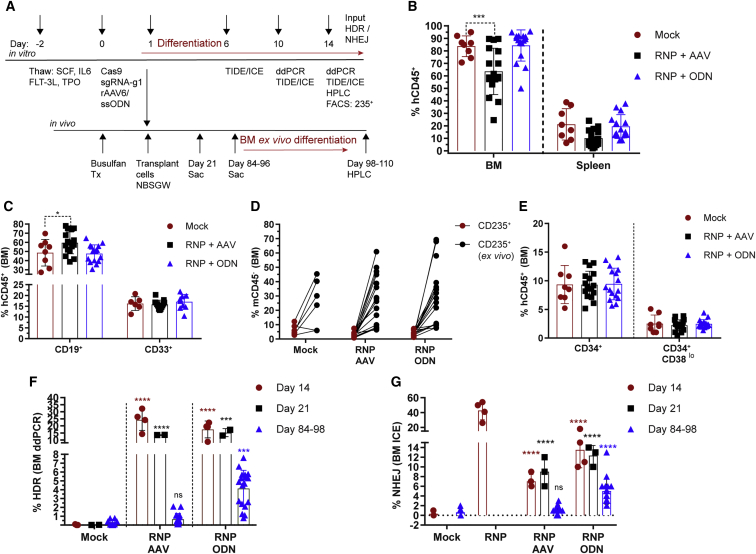
Figure 5.
Engraftment Potential of rAAV6- versus ssODN-Edited HSCs
(A) Experimental timeline for testing gene editing with GTC (E6V) rAAV6- or ssODN-treated cells in vitro in mPBSCs and in vivo in the NBSGW mouse model. Red lines indicate placement of cells in erythroid differentiation conditions. (B) Human cell (hCD45+) chimerism in the BM and spleen at days 84–96, with gating based on forward scatter (FSC), side scatter (SSC), and single cells. (C) Human CD19+ and CD33+ subsets within the BM hCD45+ population. (D) Human CD235+ cells in the BM gated on the mCD45− population. The BM cells were cultured ex vivo for 14 days in erythroid differentiation media, and CD235+ (ex vivo) was measured by flow cytometry. (E) Proportion of human CD34+ and CD34+CD38lo cells within the BM hCD45+ population. (F) HDR rates determined by ddPCR within the GTC (E6V) rAAV6- or ssODN-treated input cells (day 14, n = 4 transplants, single donor), at 3 weeks post-transplant (day 21, n = 2), and at 12–14 weeks post-transplantation (days 84–96; mock n = 8, RNP + rAAV6 n = 17, RNP + ssODN n = 18). (G) NHEJ rates determined by ICE sequencing for GTC (E6V) rAAV6- or ssODN-treated input cells (day 14), at 3 weeks (day 21) post-transplant, and at 12–14 weeks (days 84–96) post-transplant. All bar graphs show mean ± SD. ns, not significant; *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001. p value was calculated by comparing each sample mean of NHEJ (%), HDR (%), and WT (%) with the respective NHEJ (%), HDR (%), and WT (%) of the mock sample by two-way ANOVA with Dunnett’s multiple comparison. Asterisks are color matched to the respective mock sample. See also Figures S10–S15.