Figure 1.
BM450697 lncRNA Characterization in Hepatic Carcinoma Cell Lines
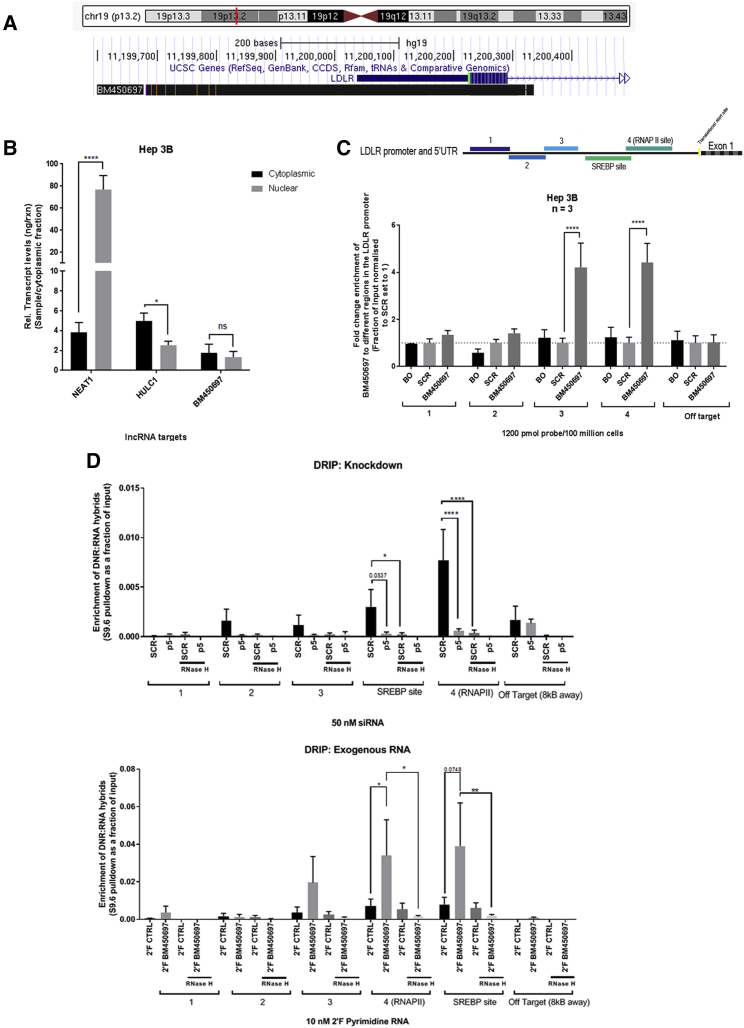
(A) A schematic diagram from the UCSC Genome Browser showing the position of BM450697 relative to the LDLR gene. (B) BM450697 appears to be equally present in the nuclear and cytoplasmic fractions. Hep 3B cells (10 million) were separated into nuclear and cytoplasmic fractions, using subcellular fractionation for RNA. HULC1 transcripts were used as positive controls for cytoplasmic expression, and NEAT1 was used as a positive control for nuclear expression. (C) BM450697 is enriched at the promoter site of LDLR in Hep 3B cells. Hep 3B cells (100 million) were harvested and incubated with either 5′ biotinylated ASOs toward BM450697 or scrambled controls, overnight at 37°C. Thereafter, a ChIRP assay was performed, the resultant genomic DNA was isolated, and subsequent qPCR was performed to determine the fold enrichment of BM450697 at the different promoter sites. BO, beads only; SCR, scrambled. (D) BM450697 formed DNA:RNA hybrids over the promoter region of the LDLR gene. Genomic DNA was isolated from either knockdown of BM450697 or exogenously added 2′ fluorinated BM450697 in Hep 3B cells after 72 h. Thereafter, 5 μg DNA was used per immunoprecipitation, in DNA samples treated with or without RNase H. Student’s t test was used in (B), comparing cytoplasmic and nuclear fractions for each gene amplified. Two-way ANOVA with the post hoc Tukey’s test was used in (C) and (D). *p < 0.05, **p < 0.01, and ***p < 0.0001.