Abstract
Despite the efficacy of BCR-ABL tyrosine kinase inhibitors (TKIs) in chronic phase-chronic myeloid leukemia, the management of blast phase-chronic myeloid leukemia (BP-CML) remains a challenge. Therefore, there is an urgent need to identify alternative agents that act synergistically with BCR-ABL TKIs in BP-CML. Our results show that the anti-malarial agent, mefloquine augments the efficacy of TKIs in CML cell lines and primary CML cells in vitro, including those with the T315I mutation. This effect is selective as mefloquine is more effective in inducing apoptosis, inhibiting colony formation and self-renewal capacity of CD34+ cells derived from TKI-resistant BP-CML patients than normal cord blood (CB) CD34+ stem/progenitor cells. Notably, the combination of mefloquine and TKIs at sublethal concentrations leads to synergistic effects in CML CD34+ cells, while sparing normal CB CD34+ cells. We further demonstrate that the initial action of mefloquine in CML cells is to increase lysosomal biogenesis and activation, followed by oxidative stress, lysosomal lipid damage and functional impairment. Taken together, our work elucidates that mefloquine selectively augments the effects of TKIs in CML stem/progenitor cells by inducing lysosomal dysfunction.
Introduction
Chronic myeloid leukemia (CML) is a hematological stem cell malignancy characterized by the reciprocal translocation of chromosomes 9 and 22, resulting in the constitutively active BCR-ABL1 tyrosine kinase. BCR-ABL1 activates a number of signal transduction pathways involved in cell survival and growth, including Ras/MEK/MAPK, PI3K/AKT, STAT and MYC [1]. Despite remarkable clinical responses achieved with BCR-ABL1 tyrosine kinase inhibitors (TKIs) in chronic phase-CML, these TKIs have been less effective as single agents in blast phase (BP) CML [2]. Mechanisms for TKI-resistance of BP-CML are complex. Apart from BCR-ABL1 overexpression and kinase mutations, increasing evidence show that CML stem/progenitor cells do not depend on BCR-ABL1 kinase activity for survival [3], [4], [5]. Hence, identification of new therapeutic targets is needed for more effective management of BP-CML.
Lysosomes are acidic organelles filled with numerous hydrolases and have been recently recognized to play an important role in inducing cell death [6]. Compared with normal cells, lysosomal function plays a more important role in cancer, as cancer progression is often characterized by dramatic changes in lysosomal volume, composition and cellular distribution [7], [8], [9]. In addition, lysosomal dysfunction has been shown to have a profound impact on cancer cell growth and survival [10], [11], suggesting that the lysosome is an attractive therapeutic target in cancer therapeutics.
Mefloquine is an anti-malarial drug used to prevent or treat malaria. Several studies have shown that mefloquine has anti-cancer properties where it induces death in tumor cells of diverse tissue origins, such as prostate, blood and breast [7], [12], [13], [14]. Mefloquine have also been found to enhance the activity of other anti-cancer drugs against tumor cells [15], [16]. Although anti-cancer mechanisms of mefloquine via ROS-mediated modulation of AMPK signaling [17] and lysosomal disruption [7] have been described, its precise molecular mechanism is still not well understood.
In this study, we investigated the effects of mefloquine alone and in combination with BCR-ABL1 TKIs using CML cell lines and primary patient CML cells, as well as cord blood (CB) samples as normal controls. We further analyzed the mechanism of the action of mefloquine in CML focusing on the lysosome. Our findings show that mefloquine preferentially targets CML CD34+ stem/progenitor cells and augments the efficacy of BCR-ABL1 TKIs by inducing lysosomal dysfunction.
Materials and Methods
Cell Lines and Reagents
Human CML cell lines, K562 (kind gift from Dr. Junia Melo), KU812 (kind gift from Dr. S Tiong Ong) and murine CML cell lines, 32Dp210 (kind gift from Dr. Brian Druker) and 32Dp210 T315I mutant (kind gift from Dr. James Griffin) were maintained in suspension in RPMI medium (Thermo Fisher Scientific, USA), supplemented with 10% fetal bovine serum, 4 mM L-glutamine (Hyclone, USA), 1% penicillin/streptomycin (Gibco, Thermo Fisher Scientific, USA). 32Dp210 and 32Dp210 T315I are murine hematopoietic 32D cells transfected with BCR-ABL1 and T315I mutant respectively [18]. The cell lines used in our study are validated with short tandem repeat (STR) profile analysis or Sanger sequencing analysis (Table S1 and Figure S1). Imatinib (LC Laboratories, USA) and ponatinib (Selleckchem, USA) were dissolved in sterile distilled water. Mefloquine hydrochloride (Sigma, US) and bafilomycin A1 (Cayman Chemicals, USA) were reconstituted in dimethylsulfoxide (DMSO; Sigma, USA). N-acetyl cysteine (NAC; Sigma, USA) was dissolved in sterile distilled water. α-Tocopherol (Sigma, USA) was dissolved in a mixture of DMSO and 30% ethanol.
Primary CML Cells
Primary CML samples were obtained from patients from the Singapore General Hospital and CB samples were obtained from the Singapore Cord Blood Bank. Written informed consent was obtained from all patients under institutional review board-approved protocols. Primary CD34+ samples are purified from mononuclear cells from peripheral blood or bone marrow samples obtained from BP-CML patients using CD34 MicroBead kit (Miltenyi Biotec, Germany). CD34+ samples with purity ≫90% (Table S2) used were cryopreserved in liquid nitrogen prior to use in our work. These samples were from patients who were in blast crisis, with corresponding mutations detected, during the time of sample collection. Isolated CD34+ cells were cultured in StemPro™-34 SFM Complete Medium (Thermo Fisher Scientific, USA), supplemented with the same cytokines as described in our previous study [19].
MTS Proliferation Assay
Cells were plated at a density of 1.0 × 104 cells per well in 96-well microplates and treated with appropriate concentrations of drugs. After 3 days, cellular proliferation was analyzed with the CellTiter 96 Aqueous One Solution Cell Proliferation assay kit (G3581, Promega, USA). To estimate the IC50 values, proliferation data obtained from MTS assays were normalized to untreated controls and graphed in Prism, with upper and lower limit capped at 100% and 0% proliferation respectively. Graphs were fit with non-linear regression fit, with use of [inhibitor] vs normalized response function in Prism 8.0.1, where IC50 were obtained through best-fit values.
Combination Index (CI) Analysis
Combination studies were performed using the same protocol as described in our previous study [19]. Briefly, the IC50 of mefloquine and TKIs (imatinib or ponatinib) was first determined in single arm experiments. Cells were then treated with increasing doses of mefloquine or appropriate TKI at an equipotent constant-ratio combination of both drugs. The cell growth of single and combination arms were determined after 48 and 72 hours. The combination index (CI) of growth inhibition was calculated using the CalcuSyn software (Biosoft, UK).
Cell Viability Assay
Cells were seeded at a density of 1 × 105 cells per well in 12-well plates and exposed to appropriate concentrations of drugs. Treated cells were stained with Annexin V-FITC and 7-aminoactinomycin D (7-AAD) (Beckman Coulter, USA) and analyzed on a Beckman Coulter FC500 flow cytometer. Percentage of Annexin V-negative/7-AAD-negative cells was determined by CXP software analysis. Viable cells were considered as Annexin V-negative/7-AAD-negative.
Colony-Forming and Serial Replating Assays
Primary CD34+ cells were plated at a density of 1–5 × 103 cells in HSC-CFU complete methylcellulose medium (Miltenyi Biotec, Germany) together with the drugs. Colonies were scored after 2 weeks to determine the colony-forming units. Subsequently, individual colonies were selected and replated in 96-well plates in HSC-CFU complete methylcellulose medium for the serial relating assay. Wells were scored after 2 weeks and recorded as positive or negative for presence of colonies. Serial replating was conducted until there was no more formation of colonies. The serial replating capacity was determined by the percentage of positive wells among total number of colonies plated.
Lysosomal pH and Activity Assays
Cells were seeded at a density of 2 × 105 in six-well plates and treated with appropriate drug concentrations. Treated cells were stained with LysoTracker™ Red DND-99, LysoSensor™ Green DND-189 (Thermo Fisher Scientific, USA), acridine orange or Magic Red Cathepsin B (ImmunoChemistry Technologies, USA) in accordance to manufacturers' protocol and then analyzed on fluorescence microscope or a MACSQuant® VYB flow cytometer.
Oxidative Stress and Damage Assays
Cells were seeded at a density of 2 × 105 in six-well plates and exposed to appropriate drug concentrations. Treated cells were stained with MitoSOX™ Red (to indicate mitochondrial superoxide) or CM-H2DCFDA (to indicate intracellular ROS, Thermo Fisher Scientific, USA) in accordance to manufacturers' protocol. Stained cells were analyzed on a MACSQuant® VYB flow cytometer. Lipid peroxidation was determined with Lipid Peroxidation (MDA) Assay Kit (ab118970, Abcam, USA) by measuring the malondialdehyde (MDA) levels.
Western Blot
Total proteins were separated electrophoretically by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) and then electroblotted onto polyvinylidene fluoride (PVDF) membranes (Bio-Rad, USA). Specific primary antibodies against LAMP1 (sc-20,011, Santa Cruz Biotechnology, USA), LAMP2 (sc-18,822, Santa Cruz Biotechnology, USA), LC3B (#3868, Cell Signaling Technology, USA), p62 (ab56416, Abcam, USA); together with appropriate HRP-linked secondary antibody were used. β-actin was used as a loading control.
Statistical Analysis
All results are indicative of at least three independent experiments. All data are expressed as mean and standard deviation. For single drug treatment and combination drug treatment, one-way and two-way ANOVA statistical analyses were conducted in Prism 8.0.1 respectively. For all ANOVA analyses, Gaussian distribution and equal SD were assumed, with no matching or pairing. For both one-way and two-way ANOVA analyses, the Dunnett's and Sidak's multiple comparisons tests were respectively done to obtain ANOVA P-values for each treatment group vs control group. For comparison between two or less treatment groups, a student's t-test was used. A P-value of ≪.05 was considered statistically significant.
Results
Mefloquine is Active Against CML Cell Lines and Synergistic with TKIs
We first investigated the effect of mefloquine on the human CML cell lines, K562 and KU812 and murine CML cell lines, 32Dp210 and 32Dp210 T315I. Using MTS proliferation assay, we found that mefloquine inhibited the proliferation of CML cell lines in a dose-dependent manner (Figure 1A and Figure S2). The IC50 for K562, KU812, 32Dp210 and 32Dp210 T315I were 10.5 μM, 8.32 μM, 8.56 μM and 7.68 μM respectively at 72 hours (Table S3). Mefloquine at 10 and 15 μM decreased CML cell viability as determined by the percentage of Annexin V/7-AAD staining (Figure 1B and Figure S3).
Figure 1.
Mefloquine inhibits proliferation and reduces cell viability in CML cells and its effects are enhanced when combined with tyrosine kinase inhibitors (TKIs). (A) Mefloquine significantly inhibits proliferation in a panel of CML cell lines: K562, KU812, 32Dp210 (wild-type) and 32Dp210 (T315I) after 24 hours. (B) Mefloquine induces loss of cell viability in all cell lines after 24 hours. Isobologram of combination indices (CI) vs fraction affected (Fa) for 32Dp210 (wild-type) (C) and 32Dp210 (T315I) (D) at combination of imatinib to mefloquine 1:100 and ponatinib to mefloquine 1:1000 respectively at 72 hours. If CI50≪ 1, the combination was synergistic; if CI50 = 1, the combination was additive; if CI50≫ 1, the combination was antagonistic. (E) K562 with imatinib (0.4 μM), KU812 with imatinib (0.3 μM), 32Dp210 (wild-type) with imatinib (0.1 μM) and 32Dp210 (T315I) with ponatinib (5 nM) exhibit further loss of cell viability when combined with mefloquine (5 μM) after 72 hours. Drugs were added simultaneously for combination studies. Data are representative of at least three independent experiments. All error bars as shown are standard deviation. *, P ≪ .05, compared to control or TKI.
To determine if the combination of mefloquine and BCR-ABL1 TKIs was synergistic, we performed combination studies based on the methods proposed by Chou and Talalay [20] and calculated the combination indices (CI). Imatinib was used in K562, KU812 and 32Dp210 whereas ponatinib was used in 32Dp210 (T315I) for combination studies. The CI of mefloquine and TKI at 50%, and 75% growth inhibition were all less than 1 in 32Dp210 and 32Dp210 (T315I) (Figure 1, C and D and Table S4) and K562 and KU812 (Figure S4, S5 and Table S4), indicating that the combination of mefloquine and TKIs is synergistic in inhibiting CML cell proliferation. Time course analysis indicated that the combination is synergistic at both 48- and 72- hour drug treatment (Figure S5). The combination of mefloquine and TKIs was also more effective in decreasing CML cell viability as compared to TKI alone (Figure 1E and Figure S6 and S7).
Mefloquine Selectively Targets BP-CML CD34+ Stem/Progenitor Cells and Acts Synergistically with TKIs
We next investigated the effect of mefloquine on CD34+ stem/progenitor cells isolated from BP-CML patients. These patients were TKI-resistant and harbored BCR-ABL1 kinase mutations, including T315I (patient clinical information in Table S5). Patients #1, #2 and #3 are CML patients harboring non-T315I mutations, while patients #4, #5 and #6 are CML patients with T315I mutation. As these patients were imatinib-resistant, we therefore used dasatinib for samples harboring non-T315I mutations and ponatinib for samples with T315I mutation in the combination studies. CB CD34+ cells were used as normal controls. Mefloquine decreased the viability of BP-CML and CB CD34+ cells in a dose-dependent manner but BP-CML CD34+ cells were more sensitive than CB CD34+ cells to mefloquine at 15 and 20 μM, suggesting a therapeutic window at these concentrations (Figure 2A and Figure S8A). Importantly, the combination of sub-toxic concentrations of mefloquine and dasatinib (for T315I-negative samples) or ponatinib (for T315I-positive samples) was significantly more effective than either single agent in decreasing the viability of BP-CML but not CB CD34+ cells (Figure 2A, Figure S8A and S9).
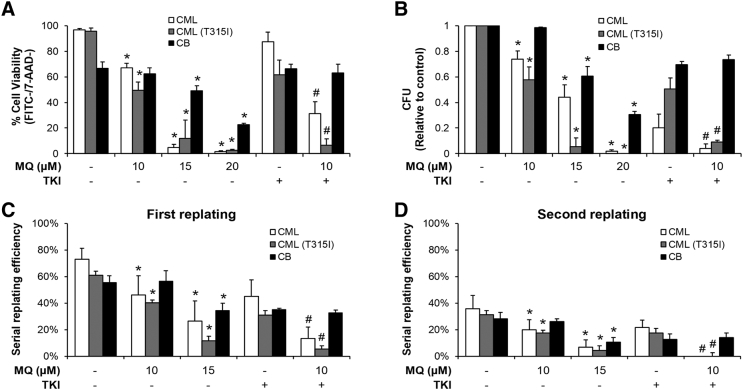
Figure 2.
Mefloquine selectively targets BP-CML CD34+ cells, with enhanced effects when combined with TKIs. (A) Mefloquine reduces cell viability in CML and CML (T315I) CD34+ cells, with enhanced effects when combined with dasatinib (100 nM) and ponatinib (5 nM) respectively. Less reduction in cell viability is observed with cord blood (CB) CD34+ cells. Cells were treated with drugs for 3 days prior to apoptosis analysis. (B) Mefloquine inhibits colony formation in CML and CML (T315I) CD34+ cells, with greater inhibition when combined with dasatinib (50 nM) and ponatinib (5 nM) respectively. Less inhibition is observed with CB CD34+ cells. Mefloquine inhibits the serial replating capacity in CML and CML (T315I) CD34+ cells, with greater inhibition when combined with dasatinib (50 nM) and ponatinib (5 nM) respectively for both the first (C) and second (D) replating. Less inhibition is observed with CB CD34+ cells. CML samples harbor other BCR-ABL1 mutations but not T315I. CML (T315I) samples harbor T315I mutation. Drugs were added simultaneously for combination studies. Data are representative of at least three independent experiments. All error bars as shown are standard deviation. *, P ≪ .05, compared to control; #, P ≪ .05, compared to TKI.
The hallmark features of CML stem/progenitor cells are proliferation, differentiation and self-renewal, which are analyzed with colony formation and serial replating assays. We found that mefloquine is more effective in inhibiting colony formation and serial replating of BP-CML CD34+ cells compared to CB CD34+ cells (Figure 2, B–D, Figure S8B to 8D and S10). Combination of mefloquine and dasatinib (for T315I-negative samples) or ponatinib (for T315I-positive samples) further inhibited colony formation and serial replating in BP-CML but not in CB CD34+ cells. Notably, the combination completely abolished the self-renewal capacity of BP-CML CD34+ cells. Taken together, mefloquine and its combination with BCR-ABL1 TKIs preferentially target the proliferative and self-renewal capacity of BP-CML CD34+ cells.
Biphasic Response in Lysosomal Activation by Mefloquine Leads to Lysosomal Dysfunction in CML Cells
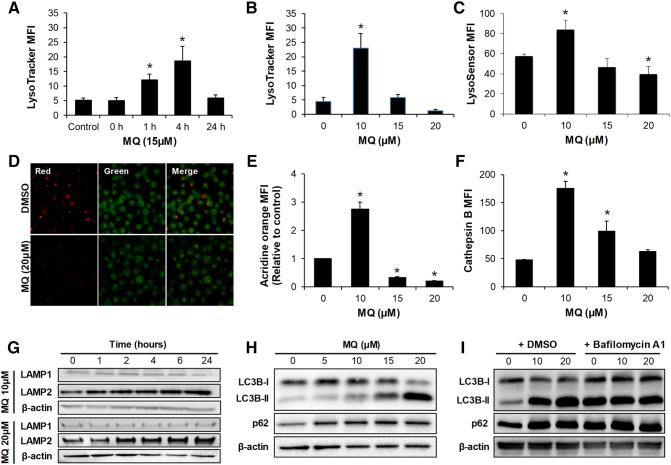
To determine whether mefloquine acts on CML via targeting the lysosome, we examined the lysosomal pH and activity in CML cells treated with mefloquine. LysoTracker, a dye that accumulates in acidic intracellular compartments, was used to assess changes in lysosomal pH [21]. We observed a time-dependent biphasic response with increased LysoTracker intensity after 1 and 4 hours of treatment with 15 μM mefloquine followed by a decrease after 24 hours (Figure 3A). These findings suggest that the initial increase in lysosomal acidification was due to a compensatory response or lysosomal adaptation to lysosomal disruption [22]. A dose-dependent biphasic response was also observed with increased LysoTracker intensity at 10 μM mefloquine but with a decrease with 15 and 20 μM (Figure 3B and Figure S11). Similar changes in lysosomal pH were assessed using acridine orange and LysoSensor stains [23]. Similar to the findings with LysoTracker dye, we observed a dose-dependent biphasic response (Figure 3, C and E). Bafilomycin A1 is a macrolide that inhibits lysosomal acidification by preventing the passage of protons into the lysosomal lumen [24]. To investigate whether lysosomal acidification blockage can rescue mefloquine-induced cell death, we performed apoptosis assay in K562 cells exposed to mefloquine and bafilomycin A1. Bafilomycin A1 partially rescued mefloquine's effect in inducing K562 cell death (Figure S12). This finding is consistent with a previous report that showed that bafilomycin A1 protects against mefloquine-mediated acute myeloid leukemia (AML) cell death [7].
Figure 3.
Mefloquine induces initial lysosomal activation as a compensatory mechanism with consequent lysosomal dysfunction in CML cells. (A) Time-dependent biphasic response with increased LysoTracker staining after 1 and 4 hours of treatment with 15 μM mefloquine followed by a decrease after 24 hours. Dose-dependent biphasic response with increased LysoTracker staining (B) and LysoSensor staining (C) at 10 μM mefloquine followed by a decrease at 15 and 20 μM. (D) Representative images showing the acidic red acridine orange staining with 20 μM mefloquine in K562 cells. (E) Dose-dependent biphasic response with increased acidic red acridine orange staining at 10 μM mefloquine followed by a decrease at 15 and 20 μM. (F) Dose-dependent biphasic response with increased Magic Red Cathepsin B staining at 10 μM mefloquine followed by a decrease at 15 and 20 μM. Cells were treated with mefloquine for 24 hours prior to Lysotracker, LysoSensor, acridine orange and Cathepsin B staining. All dyes were used at 1 μM final concentration. (G) Mefloquine at 10 μM and 20 μM induces time-dependent increase of LAMP2 but not LAMP1 protein. (H) Mefloquine induces accumulation of LC3B-II fragment and p62 after 24 hours treatment. (I) When co-treated with bafilomycin A1 (5nM) for 3 hours, there is no further increase of mefloquine-induced accumulation of LC3B-II fragment and p62. Data are representative of at least three independent experiments. All error bars as shown are standard deviation. *, P ≪ .05, compared to control.
We further assessed lysosomal activity by measuring enzyme activity of cathepsin B and expression level of lysosomal-associated membrane protein-2 (LAMP-2). We found that mefloquine increased cathepsin B activity in a dose-dependent biphasic manner and LAMP-2 levels in a time-dependent manner (Figure 3, F and G, and Figure S13A and 13B), demonstrating increased lysosomal activation. However, the lysosomal function is inhibited by mefloquine in CML cells as observed through increased LC3B-II/I ratio and p62 (Figure 3H and Figure S13, C and D). The inhibitory effect of mefloquine on lysosomal function was confirmed by our findings that mefloquine did not further increase autophagy flux in the presence of bafilomycin A (Figure 3I, Figure S13E and S13F).
Mefloquine Causes Lysosomal Lipid Damage in CML Cells
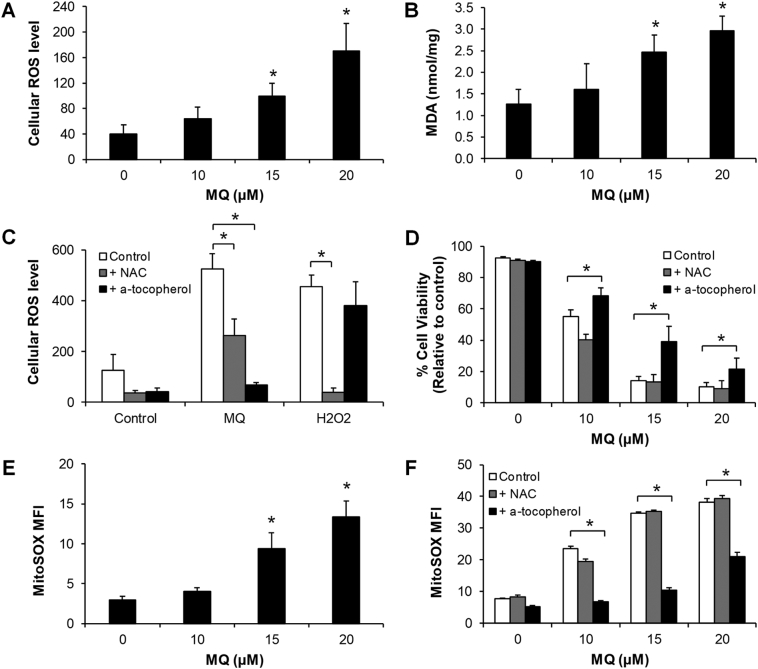
Lysosomal disruption is often associated with oxidative stress in cells [25]. In keeping with this, we found that mefloquine significantly increased ROS levels in a dose-dependent manner in CML cells after 4 hours (Figure 4A). Malondialdehyde (MDA), the end product of lipid peroxidation, was increased by mefloquine as well (Figure 4B). These findings demonstrate that mefloquine induces oxidative stress and causes oxidative damage of lipids. We also show that α-tocopherol, an antioxidant that inhibits lysosomal lipid-mediated ROS production, protects against mefloquine-induced but not H2O2-induced oxidative stress (Figure 4C), suggesting that mefloquine causes the lysosomal lipid damage. Partial rescue was observed with N-acetyl-L-cysteine (NAC), an antioxidant that inhibits thiol redox-mediated ROS production. In contrast, NAC but not α-tocopherol completely rescued H2O2-induced oxidative stress. These results demonstrate that rescue by α-tocopherol is specific to mefloquine. In addition, α-tocopherol, but not NAC, significantly reversed mefloquine's effects on CML cell viability (Figure 4D), indicating that lysosomal lipid damage is an important requisite for the efficacy of mefloquine in CML. We further show that mefloquine increases mitochondrial superoxide level (Figure 4E and Figure S14), which can be reversed by α-tocopherol but not with NAC (Figure 4F). This finding suggests an association between lysosome lipid damage and mitochondrial dysfunction. Together, our data indicate that mefloquine causes CML cell death as a consequence of lysosomal lipid damage.
Figure 4.
Mefloquine induces oxidative stress and lysosomal lipid damage. (A) Mefloquine induces dose-dependent increase of intracellular ROS levels after 4 hours treatment in K562 cells. (B) Mefloquine induces dose-dependent increase of MDA levels in K562 cells after 24 hours treatment. α-tocopherol but not NAC significantly reversed the effects of mefloquine in increasing ROS levels (C) and decreasing viability (D) in K562 cells after 24 hours. On the other hand, ROS generated by H2O2 (50 μM) could be rescued by NAC but not α-tocopherol (C). (E) Mefloquine increases MitoSOX levels in K562 cells after 24 hours treatment. (F) α-tocopherol but not NAC significantly reversed the effects of mefloquine in increasing MitoSOX levels in K562 cells after 24 hours. Antioxidants α-tocopherol and NAC were used at 10 mM. Data are representative of at least three independent experiments. All error bars as shown are standard deviation. *, P ≪ .05, compared to control.
Discussion
Lysosomes have become attractive targets for cancer therapeutics since the discovery of lysosomal adaptation in regulating cancer cell survival [8], [10], [25]. Cancer cells require increased lysosomal function to proliferate, metabolize, and adapt to stressful environments [7], [8], [9]. Particularly in leukemia, various studies using preclinical models have shown that lysosomal dysfunction is effective in eliminating leukemia bulk as well as quiescent leukemia stem cells without significant toxicity in normal counterparts in vitro and in vivo [7], [26], [27], [28]. Notably, the lysosome has been found to contribute to radiotherapy and chemotherapy resistance in cancer [29], [30]. Targeting lysosomes using Leu-Leu-OMe, a lysosomotropic agent, has been shown to eradicate imatinib-resistant CML cells [28].
Anti-cancer activity of mefloquine, a quinolone-based anti-malarial drug, has recently been demonstrated in human cancer cell lines [14], [16], [17], [31]. Although mechanisms of action of mefloquine seem to be cancer type-specific, most studies suggest that mefloquine acts on cancer cells via targeting lysosome and inhibiting autophagy [14], [7], [31]. In this work, we demonstrate that targeting lysosomes with mefloquine inhibits growth and decreases viability of BCR-ABL1 TKI-sensitive CML cells and TKI-resistant primary BP-CML cells. To the best of our knowledge, our work is the first to show the inhibitory effects of mefloquine in eliminating primary leukemic stem/progenitor cells.
Mefloquine produces a rapid and biphasic response in lysosomal biogenesis prior to inducing lysosomal lipid damage and dysfunction of CML cells in our experiments. It reflects a common feature of lysosomal response to mefloquine in mammalian cells and parasites [33], [34]. These responses are consistent with the effects of lysosomotropic agents that evoke an initial compensatory lysosomal biogenic response but with the ultimate consequence of lysosomal dysfunction [22]. Interestingly, we observe the association between lysosomal inhibition and mitochondria function, which supports the notion that lysosomes act as metabolic sensors that regulate mitochondrial metabolism [35].
Our study highlights the potential of mefloquine as a targeted therapeutic agent based on its preferential and selective elimination of CML stem/progenitor cells, in combination with BCR-ABL1 TKIs, while sparing the normal counterparts. Mefloquine significantly augments BCR-ABL1 TKIs in CML cell lines and CD34+ stem/progenitor cells, more efficiently than chloroquine, as previously described (Figure S15) [34], [35]. In contrast to mefloquine, imatinib at the same concentration used in combination studies does not affect lysosome lysotracker staining or ROS levels (Figure S16). We believe that the combinatory effects of TKIs and mefloquine observed in CML are due to the separate inhibition of BCR-ABL and lysosomal function, respectively. In healthy individuals receiving oral mefloquine 250 mg once weekly for prophylaxis of malaria, plasma concentrations of 5 μM have been observed without significant toxicities [36]. Mefloquine at a dose of 25 mg/kg has also been shown to be well tolerated in patients with moderately severe malaria with no serious adverse events [37]. A 10 μM concentration of mefloquine, shown to be highly effective in our experiments in combination with BCR-ABL1 TKIs, is likely to be clinically achievable. Notably, our work suggests that mefloquine has a greater efficacy than chloroquine (Figure S15). This is consistent with the previous work that mefloquine is more potent than chloroquine [38]. It should be noted that chloroquine and mefloquine have already been used in clinical trials for cancer (Ref NCT01430351, NCT00969306 and NCT02378532).
In conclusion, our work demonstrates that mefloquine significantly augments the effects of BCR-ABL1 TKIs in CML stem/progenitor cells while sparing normal counterparts. Our work also elucidates the cellular and molecular response to lysosomal inhibition and supports targeting lysosomes as a new therapeutic strategy in CML.
Acknowledgments
We thank Dr. S. Tiong Ong for his kind gift of the KU812 cell line, Dr. Junia V. Melo for her kind gift of the K562 cell line, Dr. Brian Druker for his kind gift of the 32Dp210 cell line, Dr. James Griffin for his kind gift of the 32Dp210 T315I cell line. We are grateful to Dr. Han Min Shen for critically reading the manuscript. This work was supported by the Singapore General Hospital Research Grants, SRG-OF#09/2016 and SRG-NIG-01 2017(W.X); National Cancer Center Singapore ONCO ACP Research Grant, NCCRF-OACPCCS-YR2015-AUG-5 (W.X); Academia Medical Research Grant AM/TP020/2018(SRDUKAMR1820) (W.X); Agency for Science, Technology and Research (A*STAR) Biomedical Research Council Translational Clinical Research Partnership Grant, BMRC/13/1/96/681 (C.C.); the National Research Foundation Singapore under its Clinician Scientist Award, NMRC/CSA/017/2010 (C.C.) and administered by the Singapore Ministry of Health's National Medical Research Council (NMRC); NMRC Centre Grant Programme -Targeted Therapy for Blood Cancer, NMRC/CG/C012A/2017 (C.C); NMRC Clinician Scientist / Clinician Investigator Salary Support Programme, NMRC/CISSP/ 2015/018a (C.C); and the Duke–NUS Medical School Signature Research Program funded by A*STAR.
Footnotes
Conflicts of Interest Statement: C.C. has received honorarium from Novartis Oncology, Bristol-Myers Squibb, Korea Otsuka Pharmaceuticals and Chiltern International, and research funding from Bristol-Myers Squibb. All other authors declare no competing financial interests.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.tranon.2019.06.001.
Contributor Information
Charles Chuah, Email: charles.chuah.t.h@singhealth.com.sg.
Wei Xiang, Email: xiang.wei@sgh.com.sg.
Appendix A. Supplementary Data
Supplementary material
References
- 1.Ren R. Mechanisms of BCR-ABL in the pathogenesis of chronic myelogenous leukaemia. Nat Rev Cancer. 2005;5:172–183. doi: 10.1038/nrc1567. [DOI] [PubMed] [Google Scholar]
- 2.Saussele S, Silver RT. Management of chronic myeloid leukemia in blast crisis. Ann Hematol. 2015;94(Suppl 2):S159–S165. doi: 10.1007/s00277-015-2324-0. [DOI] [PubMed] [Google Scholar]
- 3.Hamilton A, Helgason GV, Schemionek M, Zhang B, Myssina S, Allan EK, Nicolini FE, Muller-Tidow C, Bhatia R, Brunton VG. Chronic myeloid leukemia stem cells are not dependent on Bcr-Abl kinase activity for their survival. Blood. 2012;119:1501–1510. doi: 10.1182/blood-2010-12-326843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schmidt M, Rinke J, Schafer V, Schnittger S, Kohlmann A, Obstfelder E, Kunert C, Ziermann J, Winkelmann N, Eigendorff E. Molecular-defined clonal evolution in patients with chronic myeloid leukemia independent of the BCR-ABL status. Leukemia. 2014;28:2292–2299. doi: 10.1038/leu.2014.272. [DOI] [PubMed] [Google Scholar]
- 5.Corbin AS, Agarwal A, Loriaux M, Cortes J, Deininger MW, Druker BJ. Human chronic myeloid leukemia stem cells are insensitive to imatinib despite inhibition of BCR-ABL activity. J Clin Invest. 2011;121:396–409. doi: 10.1172/JCI35721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kroemer G, Jaattela M. Lysosomes and autophagy in cell death control. Nat Rev Cancer. 2005;5:886–897. doi: 10.1038/nrc1738. [DOI] [PubMed] [Google Scholar]
- 7.Sukhai MA, Prabha S, Hurren R, Rutledge AC, Lee AY, Sriskanthadevan S, Sun H, Wang X, Skrtic M, Seneviratne A. Lysosomal disruption preferentially targets acute myeloid leukemia cells and progenitors. J Clin Invest. 2013;123:315–328. doi: 10.1172/JCI64180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kallunki T, Olsen OD, Jaattela M. Cancer-associated lysosomal changes: friends or foes? Oncogene. 2013;32:1995–2004. doi: 10.1038/onc.2012.292. [DOI] [PubMed] [Google Scholar]
- 9.Colombo F, Trombetta E, Cetrangolo P, Maggioni M, Razini P, De Santis F, Torrente Y, Prati D, Torresani E, Porretti L. Giant Lysosomes as a Chemotherapy Resistance Mechanism in Hepatocellular Carcinoma Cells. PLoS One. 2014;9 doi: 10.1371/journal.pone.0114787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gyparaki MT, Papavassiliou AG. Lysosome: the cell's 'suicidal bag' as a promising cancer target. Trends Mol Med. 2014;20:239–241. doi: 10.1016/j.molmed.2014.01.009. [DOI] [PubMed] [Google Scholar]
- 11.Coutinho MF, Alves S. From rare to common and back again: 60years of lysosomal dysfunction. Mol Genet Metab. 2016;117:53–65. doi: 10.1016/j.ymgme.2015.08.008. [DOI] [PubMed] [Google Scholar]
- 12.Yan KH, Lin YW, Hsiao CH, Wen YC, Lin KH, Liu CC, Hsieh MC, Yao CJ, Yan MD, Lai GM. Mefloquine induces cell death in prostate cancer cells and provides a potential novel treatment strategy in vivo. Oncol Lett. 2013;5:1567–1571. doi: 10.3892/ol.2013.1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kim JH, Choi AR, Kim YK, Yoon S. Co-treatment with the anti-malarial drugs mefloquine and primaquine highly sensitizes drug-resistant cancer cells by increasing P-gp inhibition. Biochem Biophys Res Commun. 2013;441:655–660. doi: 10.1016/j.bbrc.2013.10.095. [DOI] [PubMed] [Google Scholar]
- 14.Sharma N, Thomas S, Golden EB, Hofman FM, Chen TC, Petasis NA, Schonthal AH, Louie SG. Inhibition of autophagy and induction of breast cancer cell death by mefloquine, an antimalarial agent. Cancer Lett. 2012;326:143–154. doi: 10.1016/j.canlet.2012.07.029. [DOI] [PubMed] [Google Scholar]
- 15.Fujita R, Ishikawa M, Takayanagi M, Takayanagi Y, Sasaki K. Enhancement of doxorubicin activity in multidrug-resistant cells by mefloquine. Methods Find Exp Clin Pharmacol. 2000;22:281–284. doi: 10.1358/mf.2000.22.5.796646. [DOI] [PubMed] [Google Scholar]
- 16.Li H, Jiao S, Li X, Banu H, Hamal S, Wang X. Therapeutic effects of antibiotic drug mefloquine against cervical cancer through impairing mitochondrial function and inhibiting mTOR pathway. Can J Physiol Pharmacol. 2017;95:43–50. doi: 10.1139/cjpp-2016-0124. [DOI] [PubMed] [Google Scholar]
- 17.Yan KH, Yao CJ, Hsiao CH, Lin KH, Lin YW, Wen YC, Liu CC, Yan MD, Chuang SE, Lai GM. Mefloquine exerts anticancer activity in prostate cancer cells via ROS-mediated modulation of Akt, ERK, JNK and AMPK signaling. Oncol Lett. 2013;5:1541–1545. doi: 10.3892/ol.2013.1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Matulonis U, Salgia R, Okuda K, Druker B, Griffin JD. Interleukin-3 and p210 BCR/ABL activate both unique and overlapping pathways of signal transduction in a factor-dependent myeloid cell line. Exp Hematol. 1993;21:1460–1466. [PubMed] [Google Scholar]
- 19.Xiang W, Cheong JK, Ang SH, Teo B, Xu P, Asari K, Sun WT, Than H, Bunte RM, Virshup DM. Pyrvinium selectively targets blast phase-chronic myeloid leukemia through inhibition of mitochondrial respiration. Oncotarget. 2015;6:33769–33780. doi: 10.18632/oncotarget.5615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chou TC. Drug combination studies and their synergy quantification using the Chou-Talalay method. Cancer Res. 2010;70:440–446. doi: 10.1158/0008-5472.CAN-09-1947. [DOI] [PubMed] [Google Scholar]
- 21.Lemieux B, Percival MD, Falgueyret JP. Quantitation of the lysosomotropic character of cationic amphiphilic drugs using the fluorescent basic amine Red DND-99. Anal Biochem. 2004;327:247–251. doi: 10.1016/j.ab.2004.01.010. [DOI] [PubMed] [Google Scholar]
- 22.Lu S, Sung T, Lin N, Abraham RT, Jessen BA. Lysosomal adaptation: How cells respond to lysosomotropic compounds. PLoS One. 2017;12 doi: 10.1371/journal.pone.0173771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pierzynska-Mach A, Janowski PA, Dobrucki JW. Evaluation of acridine orange, LysoTracker Red, and quinacrine as fluorescent probes for long-term tracking of acidic vesicles. Cytometry A. 2014;85:729–737. doi: 10.1002/cyto.a.22495. [DOI] [PubMed] [Google Scholar]
- 24.Mauvezin C, Neufeld TP. Bafilomycin A1 disrupts autophagic flux by inhibiting both V-ATPase-dependent acidification and Ca-P60A/SERCA-dependent autophagosome-lysosome fusion. Autophagy. 2015;11:1437–1438. doi: 10.1080/15548627.2015.1066957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dielschneider RF, Henson ES, Gibson SB. Lysosomes as Oxidative Targets for Cancer Therapy. Oxidative Med Cell Longev. 2017;2017:3749157. doi: 10.1155/2017/3749157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Baquero P, Dawson A, Mukhopadhyay A, Kuntz EM, Mitchell R, Olivares O, Ianniciello A, Scott MT, Dunn K, Nicastri MC. Targeting quiescent leukemic stem cells using second generation autophagy inhibitors. Leukemia. 2019;33:981–994. doi: 10.1038/s41375-018-0252-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Das S, Dielschneider R, Chanas-LaRue A, Johnston JB, Gibson SB. Antimalarial drugs trigger lysosome-mediated cell death in chronic lymphocytic leukemia (CLL) cells. Leuk Res. 2018;70:79–86. doi: 10.1016/j.leukres.2018.06.005. [DOI] [PubMed] [Google Scholar]
- 28.Dielschneider RF, Eisenstat H, Mi S, Curtis JM, Xiao W, Johnston JB, Gibson SB. Lysosomotropic agents selectively target chronic lymphocytic leukemia cells due to altered sphingolipid metabolism. Leukemia. 2016;30:1290–1300. doi: 10.1038/leu.2016.4. [DOI] [PubMed] [Google Scholar]
- 29.Zhang X, Wang J, Li X, Wang D. Lysosomes contribute to radioresistance in cancer. Cancer Lett. 2018;439:39–46. doi: 10.1016/j.canlet.2018.08.029. [DOI] [PubMed] [Google Scholar]
- 30.Kolb-Lenz D, Fuchs R, Lohberger B, Heitzer E, Meditz K, Pernitsch D, Pritz E, Groselj-Strele A, Leithner A, Liegl-Atzwanger B. Characterization of the endolysosomal system in human chordoma cell lines: is there a role of lysosomes in chemoresistance of this rare bone tumor? Histochem Cell Biol. 2018;150:83–92. doi: 10.1007/s00418-018-1673-x. [DOI] [PubMed] [Google Scholar]
- 31.Shin JH, Park SJ, Jo YK, Kim ES, Kang H, Park JH, Lee EH, Cho DH. Suppression of autophagy exacerbates Mefloquine-mediated cell death. Neurosci Lett. 2012;515:162–167. doi: 10.1016/j.neulet.2012.03.040. [DOI] [PubMed] [Google Scholar]
- 33.Go ML, Lee HS. Effects of mefloquine on the release of marker enzymes from the rat liver crude lysosomal fraction. Jpn J Pharmacol. 1990;53:195–199. doi: 10.1254/jjp.53.195. [DOI] [PubMed] [Google Scholar]
- 34.Paivandy A, Calounova G, Zarnegar B, Ohrvik H, Melo FR, Pejler G. Mefloquine, an anti-malaria agent, causes reactive oxygen species-dependent cell death in mast cells via a secretory granule-mediated pathway. Pharmacol Res Perspect. 2014;2 doi: 10.1002/prp2.66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Todkar K, Ilamathi HS, Germain M. Mitochondria and Lysosomes: Discovering Bonds. Frontiers in cell and developmental biology. 2017;5:106. doi: 10.3389/fcell.2017.00106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Charles BG, Blomgren A, Nasveld PE, Kitchener SJ, Jensen A, Gregory RM, Robertson B, Harris IE, Reid MP, Edstein MD. Population pharmacokinetics of mefloquine in military personnel for prophylaxis against malaria infection during field deployment. Eur J Clin Pharmacol. 2007;63:271–278. doi: 10.1007/s00228-006-0247-3. [DOI] [PubMed] [Google Scholar]
- 37.Basano SA, Bianco A, Taylor WR, Olliaro P, Camargo LM. An in vivo test to assess mefloquine 25 mg/kg for the treatment of uncomplicated falciparum malaria in Rondonia, Brazil, The Brazilian journal of infectious diseases: an official publication of the Brazilian Society of Infectious Diseases. 2006;10:279–282. doi: 10.1590/s1413-86702006000400013. [DOI] [PubMed] [Google Scholar]
- 38.Golden EB, Cho HY, Hofman FM, Louie SG, Schonthal AH, Chen TC. Quinoline-based antimalarial drugs: a novel class of autophagy inhibitors. Neurosurg Focus. 2015;38:E12. doi: 10.3171/2014.12.FOCUS14748. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material