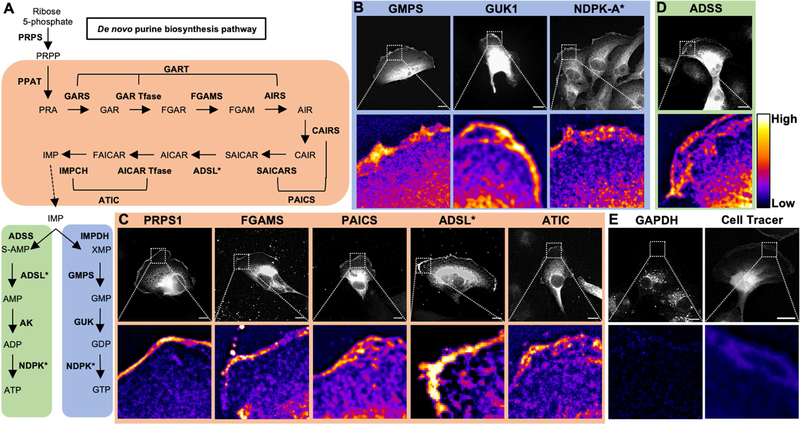
Figure 2: Nucleotide metabolic enzymes localize at the lamellipodial membrane.
Purine nucleotide metabolism pathway. De novo enzymes (orange), ATP biosynthetic branch (green), and GTP biosynthetic branch (blue) (A). Fluorescence imaging for GTP biosynthetic enzymes GMPS (mCherry-tagged, 786-o), GUK1 (Myc, Caki-1), and NDPK-A (antibody, Caki-1) (B). Immunofluorescence staining for de novo enzymes PRPS1 (Myc, Caki-1), FGAMS (GFP, Caki-1), PAICS (GFP, Caki-1), ADSL (GFP, 786-o), and ATIC (GFP, Caki-1) (C). Insets of leading edge in pseudocolor shown below. Scale bars indicate 10 μm. Asterisk (*) indicates enzymes involved in multiple pathways. PRPS: phosphoribosyl pyrophosphate synthetase. PPAT: phosphoribosyl pyrophosphate amidotransferase. GART: phosphoribosylglycinamide formyltransferase. FGAMS: phosphoribosyl formylglycinamidine synthase. PAICS: phosphoribosyl aminoimidazole succinocarboxamide synthetase. ADSL: adenylosuccinate lyase. ATIC: 5-amino-4-imidazolecarboxamide ribonucleotide transformylase/IMP cyclohydrolase. ADSS: adenylosuccinate synthase. AK: adenylate kinase. IMPDH: inosine-5′-monophosphate dehydrogenase. GMPS: GMP synthase. GUK1: guanylate kinase 1. NDPK: nucleoside-diphosphate kinase. GAPDH: glyceraldehyde 3-phosphate dehydrogenase.