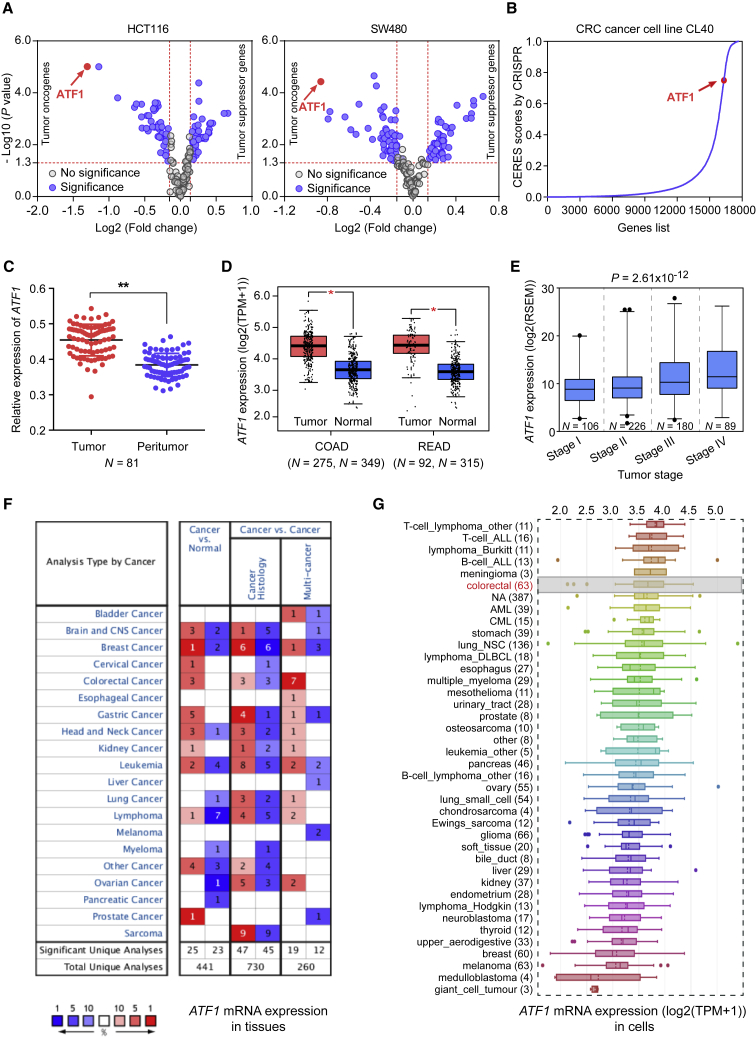
Figure 1.
Functional Genomic Screening Reveals that ATF1 Is an Oncogene in CRC
(A) Functional genomic screening based on high-throughput RNAi interrogation was used to identify genes essential for cell proliferation in the CRC risk loci in Asian populations in HCT116 and SW480 cells. Both p < 0.05 and an n- fold change >1.1 or <0.9 were selected as the threshold of significance and calculated by a two-sided Student’s t test.
(B) According to the data of a genome-wide CRISPR-Cas9-based loss-of-function screen, ATF1 is essential for cell growth; higher CERES scores are found in CRC CL40 cells. Higher CERES scores demonstrate an elevated dependency of cell viability on given genes.
(C and D) ATF1 is significantly overexpressed in tumors compared to in the normal tissues from our CRC patients and the TCGA and GTEX datasets. Data were shown as the mean ± SD and all ∗p < 0.05 and ∗∗p < 0.01 values were calculated by a two-sided Student’s t test. Abbreviations are as follows: COAD = colon adenocarcinoma and READ = rectal adenocarcinoma.
(E) ATF1 expression levels were measured in different tumor stages of CRC. Data were presented as the mean ± SD from the TCGA database. P values were calculated by one-way ANOVA.
(F and G) ATF1 expression levels were evaluated in multiple tumor tissue types from the Oncomine database and in 1,036 human cancer cell lines from the CCLE database.