Figure 4.
Transcription Factors SP1 and GATA3 Correlate with ATF1 Expression in an Allele-Specific Manner
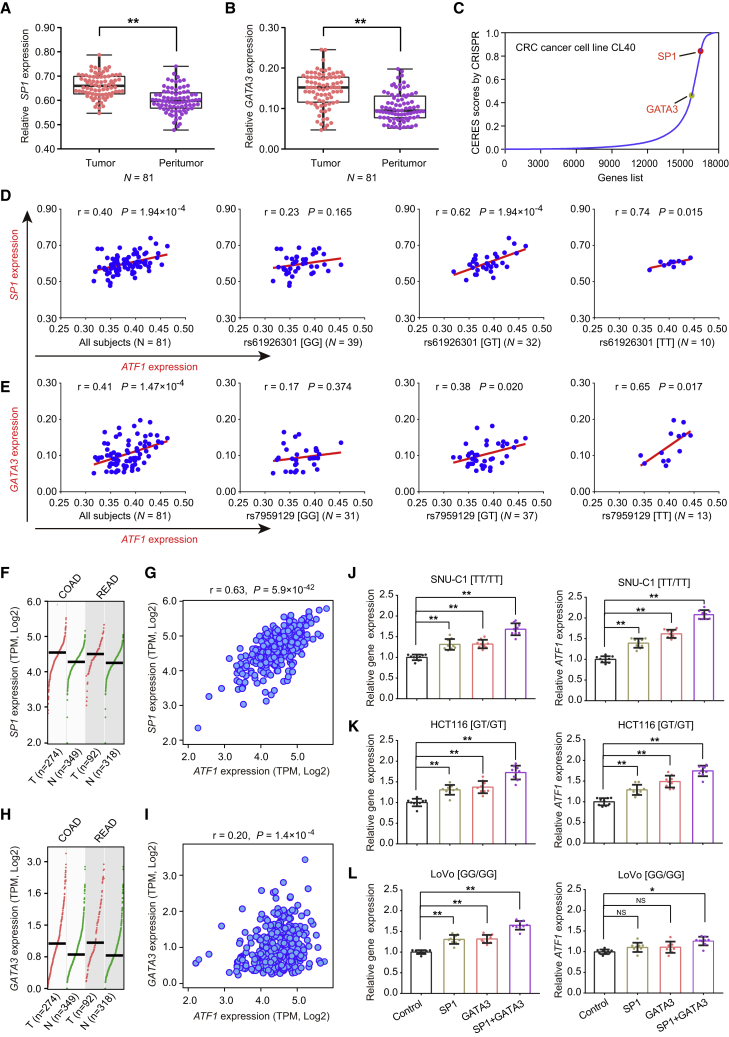
(A and B) SP1 (A) and GATA3 (B) are significantly overexpressed in tumors compared with the adjacent normal tissues from our CRC patients. Data were shown as the mean ± SD, and ∗∗p < 0.01 values were calculated with two-sided paired Student’s t test.
(C) SP1 and GATA3 are essential for cell growth; higher CERES scores are found in the CRC CL40 cells from the genome-wide CRISPR-Cas9-based loss-of-function screen data.
(D and E) The correlations of SP1 and GATA3 expression with ATF1 expression were measured in our CRC patients. All P values and r values were calculated with Spearman’s correlation analysis.
(F and G) SP1 (F) and GATA3 (G) are significantly overexpressed in tumors compared to adjacent normal tissues from the TCGA and GTEX datasets. Data were shown as the mean ± SD, and all ∗p < 0.05 values were calculated with a two-sided Student’s t test.
(H–L) The correlations of SP1 and GATA3 expression and ATF1 expression were calculated in TCGA CRC samples (H and I) and CRC cells (SNU-C1 [J], HCT116 [K], and LoVo [L]) with different genotypes of dbSNP: rs61926301 and dbSNP: rs7959129. ∗p < 0.05 and ∗∗p < 0.01; all p and r values in Figures 4H and 4I were calculated by Spearman’s correlation analysis, and all p values in Figures 4J–4L were calculated by a two-sided Student’s t test. Notably, in Figures 4J–4L, the left panels show expression of these genes (SP1, GATA3, and SP1+GATA3) in the x axis, and the right panels show the expression of ATF1.