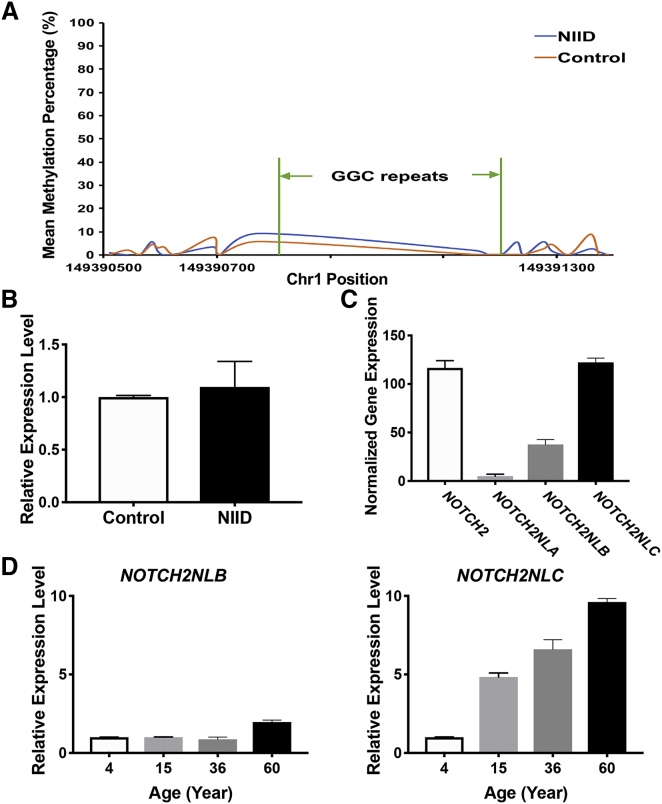
Figure 4.
Methylation and Expression at NOTCH2NLC Locus
(A) Methylation status across expanded GGC repeats region was determined using LRS data from seven affected individuals (F1-IV:7, F1-IV:15, F2-II:3, F4-II:2, F5-II:1, F5-II:4, F9-II:6) and three healthy control subjects, and no significant methylation difference was detected between NIID-affected case subjects and control subjects. Wald test was performed for statistical analysis; ∗p < 0.05.
(B) NOTCH2NLC expression level in both NIID-affected case subjects (NIID) and normal control subjects (Control). The ezDNase-treated total RNA isolated from the blood of both NIID-affected case subjects and control subjects was reversely transcribed into cDNA followed by quantitative PCR. GAPDH was used as internal control. Error bars represent the SD; Student’s t test was performed for statistical analysis; ∗p < 0.05. p = 0.776 (ns).
(C) Expression levels of NOTCH2 and three NOTCH2NL paralogs (NOTCH2NLA, NOTCH2NLB, and NOTCH2NLC) in human adult cortex detected by RNA-seq. Significant differences were observed in the expression levels of these four genes. Shown are the normalized gene expression levels. Error bars represent the SD.
(D) Dynamic change of NOTCH2NLC expression in human brain during aging. Relative expression levels of both NOTCH2NLB (left) and NOTCH2NLC (right) in DLPFC region of human brain during aging are shown. The total RNA isolated from the DLPFC of human postmortem brains from 4-, 15-, 36-, and 60-year-old subjects were used for quantitative RT-PCR with GAPDH as internal control. Error bars represent the SD.