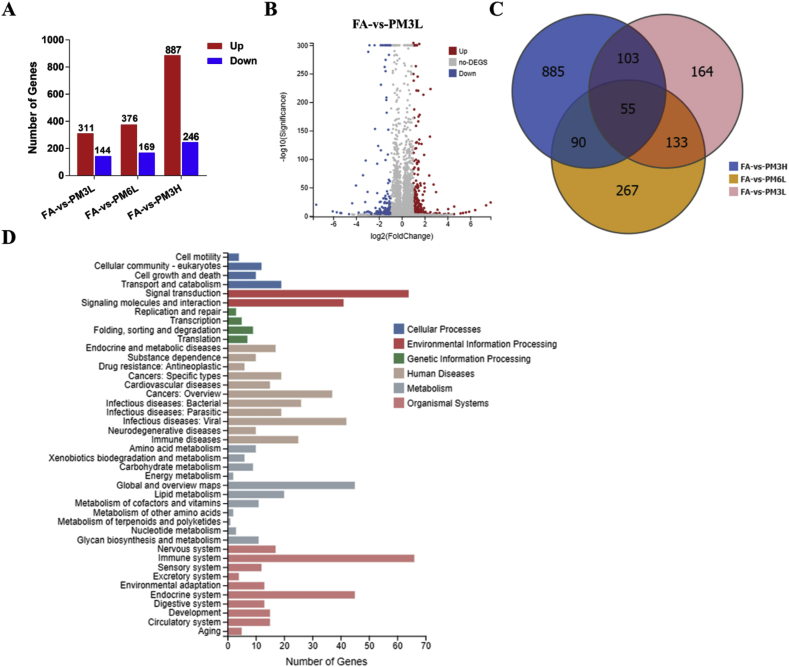
Fig. 3.
Analysis of differentially expressed genes between FA and PM2.5-exposed mice lungs. (A) Summery of the number of up- and down-regulated genes in the PM2.5-3L-, PM2.5-6L-, and PM2.5-3H-exposed lungs versus FA-exposed lung. Fold change ≥ 2 and adjusted p value ≤ 0.001 were used as the threshold to judge the significance of gene expression difference. (B) The fold change of differentially expressed genes (DEGs) of FA vs PM2.5–3L were visualized by Volcano plot. (C) The number of different and overlapped DEGs from FA vs PM2.5–3L, FA vs PM2.5–6L and FA vs PM2.5–3H group were illustrated in Venn diagram. (D) KEGG classification of DEGs from FA vs PM2.5–3L group. The functions of genes identified cover six main categories: cellular processes, environmental information processing, genetic information processing, human disease, metabolism and organismal system.