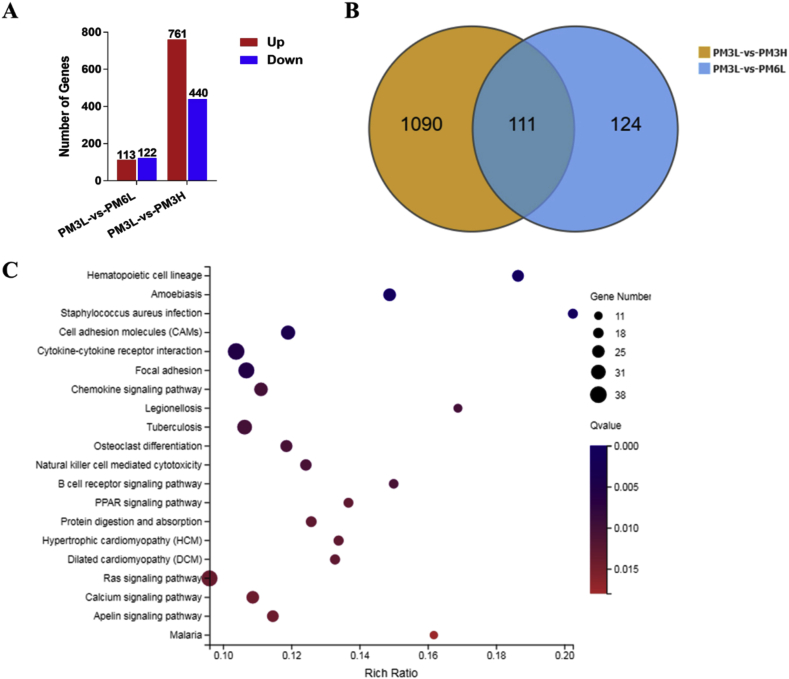
Fig. 4.
Effect of exposure time and concentration on gene expression profile in PM2.5-exposed lungs. (A) Summery of the number of up- and down-regulated genes in the PM2.5-6L-, and PM2.5-3H-exposed lungs vs PM2.5–3L -exposed lung. Fold change ≥ 2 and adjusted p value ≤ 0.001 were used as the threshold to judge the significance of gene expression difference. (B) Venn diagram shows the number of different and overlapped DEGs from PM2.5–3L vs PM2.5–6L and PM2.5–3L vs PM2.5–3H group. (C) Advanced bubble chart shows the top 20 significantly enriched KEGG pathways of DEGs in PM2.5–3L vs PM2.5–3H group. Y-axis label represents pathway, and X-axis label represents rich ratio. Size and color of the bubble represent amount of DEGs enriched in pathway and enrichment significance, respectively. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)