Figure 5.
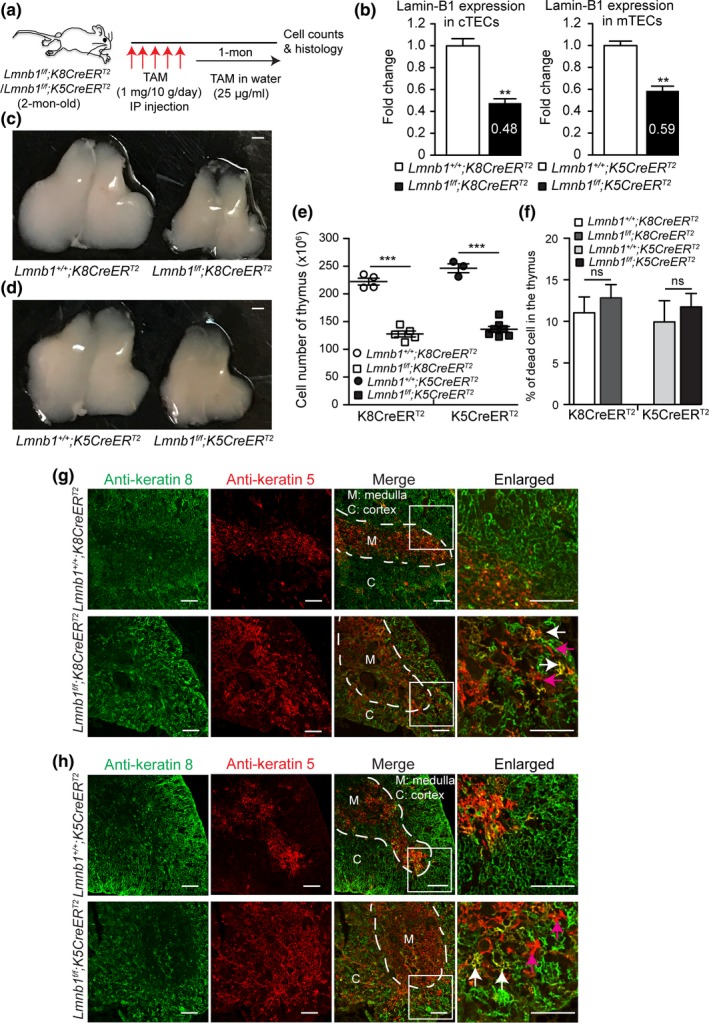
Effects of postnatal lamin‐B1 deficiency in TECs on thymus size and organization. (a) The tamoxifen (TAM)‐mediated Lmnb1 deletion scheme. Keratin 8 (K8)‐ or keratin 5 (K5)‐driven CreERT2 was used to delete Lmnb1 in cTECs or mTECs, respectively, by injecting TAM followed by feeding in TAM‐containing drinking water before analyses. (b) RT–qPCR analyses of lamin‐B1 mRNA levels revealing the Lmnb1deletion efficiency by K8CreERT2 in cTECs (left) or by K5CreERT2 in mTECs (right). The average mRNA levels of lamin‐B1 were calculated from 3 independent experiments with the control set to 1. (c,d) Representative images of thymuses from mice with indicated genotypes after TAM treatment as shown in (a). Scale bars, 1 mm. (e) The total thymic cells were counted and plotted for indicated genotypes as shown. (f) Summary of cell death in the thymus from mice with the indicated ages and genotypes. The dead cells were identified as DAPI+ populations by flow cytometry. (g,h) Keratin staining of thymuses from mice with indicated genotypes treated by TAM as shown in (a). White dash lines demarcate the cortical and medulla junction (CMJ) regions in the thymus. A section (white squares) of the thymus in each genotype was enlarged to show the well‐defined (in the controls) or disorganized (in the mutants) CMJ. Arrows mark the spreading of K5+K8+ TECs (white) and K5+ TECs (purple) into the cortical regions in the in Lmn1f/f;K8CreERT2 (g) or Lmn1f/f;K5CreERT2 (h) thymuses. Scale bars, 100 μm. Error bars, SEM from ≥ 3 independent experiments. Student's t test: *p < 0.05, **p < 0.01, ***p < 0.001, ns: not significant
