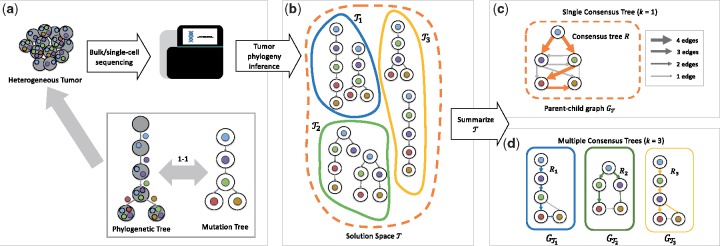
Fig. 1.
(a) Tumors are heterogeneous, composed of multiple clones with different sets of somatic mutations. This heterogeneity is the result of an evolutionary process, as modeled by a phylogenetic tree. Under the commonly used infinite sites model of evolution, where each mutation is acquired once and never lost, a phylogenetic tree may be equivalently represented by a mutation tree. (b) Due to ambiguities in bulk and single-cell sequencing data of tumors, current methods infer a large solution space of plausible mutation trees . For further downstream analyses of tumorigenesis, this solution space needs to be summarized. (c) Current summary methods either construct the parent-child graph GT or identify a single consensus tree R, failing to adequately summarize solution spaces comprised of clusters of trees with distinct topological features. (d) Here, we introduce the Multiple Consensus Tree problem to simultaneously cluster mutation trees and construct a consensus tree of each cluster