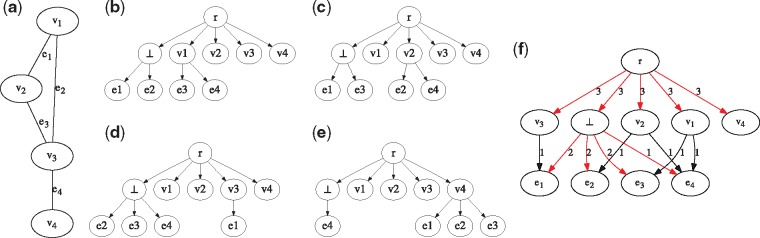
Fig. 2.
An example reduction from the Clique problem to MCT. (a) An undirected graph H with edges and vertices, containing a clique of size 3. (b–e) The n = 4 input trees to the MCT problem obtained from H. The problem instance of determining whether H contains a clique of size c = 3 reduces to the MCT instance where . An optimal clustering σ for yields and . (f) The parent-child graph , with the optimal consensus tree R1 for input trees indicated in red. The parent-child graph of is identical to T4 with edge weights for each edge (u, v) such that the corresponding optimal consensus tree R2 equals T4. As such, the total distance equals . By Lemma 6, H contains a clique of size c = 3