Fig. 8.
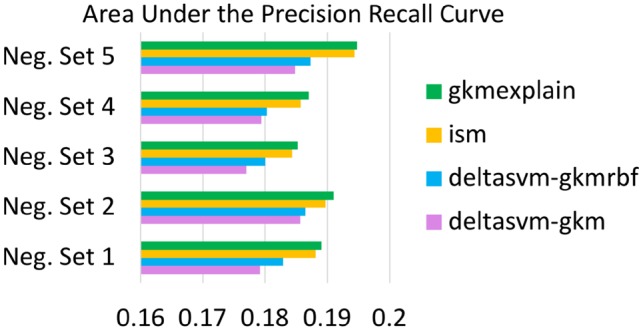
GkmExplain Mutation Impact Scores Outperform deltaSVM and ISM at identifying dsQTLs. For each choice of negative set provided in the deltaSVM paper, we trained a gkm-SVM and gkmrbf-SVM. LCL dsQTLs and control SNPs were then scored using four methods: deltaSVM on the gkm-SVM, deltaSVM on the gkmrbf-SVM, ISM on the gkmrbf-SVM and GkmExplain Mutation Impact Scores (Section 5.2) on the gkmrbf-SVM. For ISM and GkmExplain, a 51 bp window centered around the SNP was used as context. The GkmExplain mutation impact scores consistently produce the best auPRC across all 5 choices of the negative set (binomial P-value = ). SHAP was excluded from the comparison due to a prohibitively large runtime
