Fig. 2.
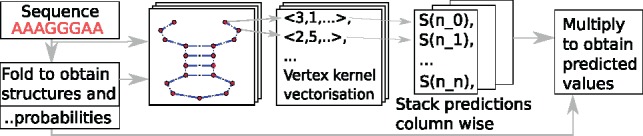
SHAPE reactivity prediction. (left) For a given sequence, possible structures are sampled. Respective Boltzmann probabilities form the unit probability vector π which has as many entries as there were structures sampled. (middle) RNA-graphs of the structures are vectorized and we use the model to predict a SHAPE (S) value for each nucleotide (n_x). We stack the predictions for each graph column wise. (right) We obtain as many predictions for every nucleotide in the sequence as there are sampled structures. We weight these by the probabilities of the structure to obtain the final prediction i.e. we multiply the matrix of predictions with π
