Fig. 3.
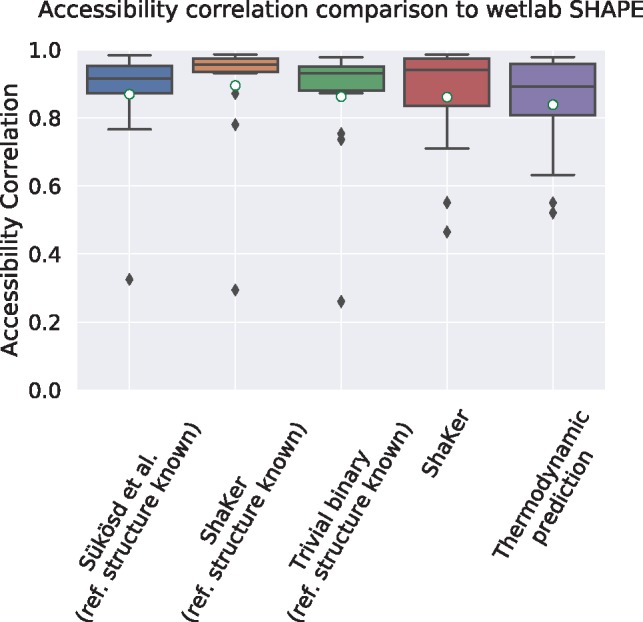
Correlation of accessibility profiles by RNAplfold using predicted SHAPE reactivities compared to the accessibility profile generated from experimental SHAPE data. The first three prediction methods are using a manually curated reference structure. Because of this prior information, all prediction tools result in a high correlation to the profile generated from experimental SHAPE data. The final two plots evaluate the situation for the more realistic application scenario where no reference structure is given. In this case, only ShaKer without structure and RNAplfold without SHAPE data can be compared. The predicted SHAPE data by ShaKer leads to an improved result compared to the pure thermodynamic prediction (mean 0.861±0.17 versus 0.839±0.16)
