Fig. 1.
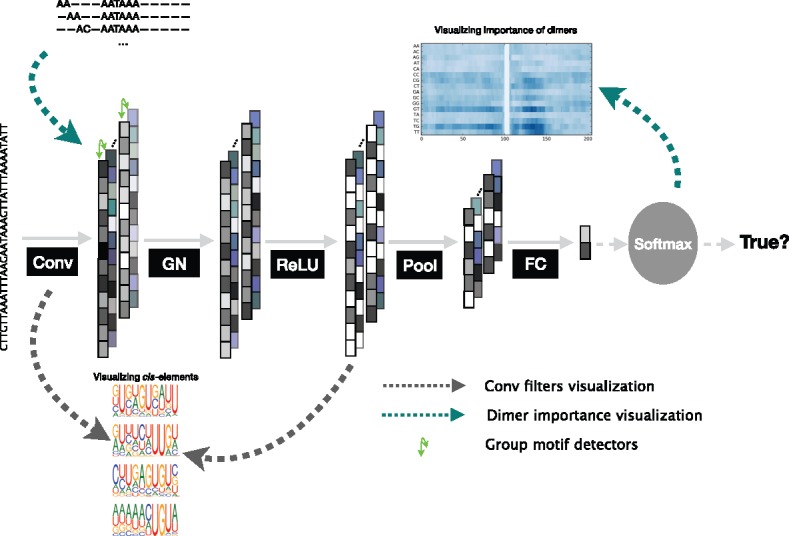
The architecture of the proposed DeeReCT-PolyA network. The output feature channels (shown as a column) of the conv layer is divided into groups (green arrows) and each group is jointly normalized by the group normalization layer. After tunable parameters are learned from the data, two visualization methods (shown as dashed lines in green and gray) are applied to the model without normalization to extract cis-elements and variants for the regulation of polyadenylation (Color version of this figure is available at Bioinformatics online.)
