Abstract
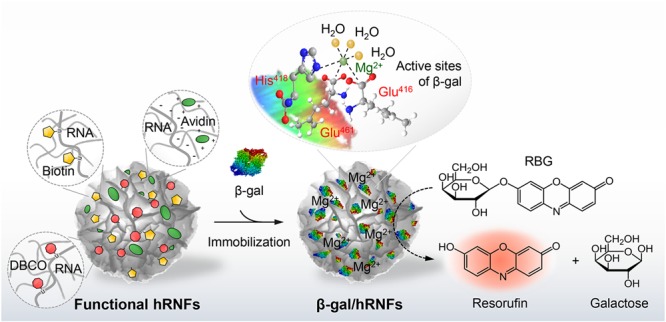
Programmable nucleic acids have emerged as powerful building blocks for the bottom-up fabrication of two- or three-dimensional nano- and microsized constructs. Here we describe the construction of organic–inorganic hybrid RNA flowers (hRNFs) via rolling circle transcription (RCT), an enzyme-catalyzed nucleic acid amplification reaction. These hRNFs are highly adaptive structures with controlled sizes, specific nucleic acid sequences, and a highly porous nature. We demonstrated that hRNFs are applicable as potential biological platforms, where the hRNF scaffold can be engineered for versatile surface functionalization and the inorganic component (magnesium ions) can serve as an enzyme cofactor. For surface functionalization, we proposed robust and straightforward approaches including in situ synthesis of functional hRNFs and postfunctionalization of hRNFs that enable facile conjugation with various biomolecules and nanomaterials (i.e., proteins, enzymes, organic dyes, inorganic nanoparticles) using selective chemistries (i.e., avidin–biotin interaction, copper-free click reaction). In particular, we showed that hRNFs can serve as soft scaffolds for β-galactosidase immobilization and greatly enhance enzymatic activity and stability. Therefore, the proposed concepts and methodologies are not only fundamentally interesting when designing RNA scaffolds or RNA bionanomaterials assembled with enzymes but also have significant implications on their future utilization in biomedical applications ranging from enzyme cascades to biosensing and drug delivery.
Keywords: rolling circle transcription, RNA flowers, organic−inorganic hybrid structures, enzyme immobilization, enzymatic study, allosteric effect
1. Introduction
Nanoscale engineering has advanced the fabrication and application of materials, with transformative impacts in a number of scientific fields. Different types of patterned organizations and highly ordered nanoscale structures have been constructed through either top-down or bottom-up approaches. In particular, nucleic-acid-based nanomaterials have emerged as powerful building blocks for the controlled bottom-up fabrication of highly structured two- or three-dimensional nano- and micrometer-sized constructs.1
Nucleic acid-based nanostructures possess advantageous properties such as sequence-driven programmability, nanoscale addressability of the created objects, and versatile bioconjugation strategies with other molecules. The DNA origami field, for example, utilizes self-assembled DNA constructs with nanometer precision built on the basis of nucleic acid hybridization. These sophisticated nanostructures can serve as excellent scaffolds to immobilize biomolecules and have been shown to effectively enhance catalytic activity2,3 and enzyme stability.2 Particularly, the precise localization of enzyme cascades on DNA origami scaffolds4 and the resulting enhancement of the cascade throughput have generated much excitement. Various types of DNA origamis5 have been created and shown to provide a programmable tool not only for the spatial organization of enzymes at specific sites, but also for the regulation of their activities on the nanoscale.6−8 However, the synthesis of DNA origami requires hundreds of unique oligonucleotides and suffers from the shortcomings of multiple annealing steps, complicated purification procedures, and small-scale production.9 Therefore, of great importance is the concern of how the assembly of nucleic acid nanostructures and further conjugation with biomolecules could be achieved in a robust and cost-effective manner.
RNA nanotechnology resembles the characteristic features of DNA-based architectures built via canonical base pairing, while providing structural flexibility, functional diversity, and thermal stability over DNA nanoparticles.10,11 RNA commonly forms well-tailored, relatively stable, and complex three-dimensional structures by adopting such properties including canonical or noncanonical base pairing, base stacking, and secondary or tertiary structural motifs. This is typically found in many biological RNAs and found less often in DNAs. Functional RNAs such as RNA aptamers, ribozymes, riboswitches, siRNA, miRNA, and other protein-mimicking or noncoding RNA molecules can be simply fused or hybridized into RNA constructs, adding multiple functionalities to the tailored RNA nanostructures. Thus, RNA architectures would provide a promising direction for creation of multifunctional RNA scaffolds with thermodynamic stability compared to their DNA counterparts. Despite their potential, RNA materials possess limitations such as difficulties in mass production and high cost, similar to limitations for DNA origamis. With regard to this, isothermal nucleic acid amplification techniques as an alternative method have been recently explored to construct soft scaffolds for many biomolecules.12
Rolling circle replication (RCR)-based nucleic acid amplification,13 including rolling circle amplification (RCA) and rolling circle transcription (RCT), has attracted tremendous interest as a powerful tool for large-scale nucleic acid production. In RCR, DNA or RNA polymerases cyclically navigate a small circular template DNA (typically 25–100 nucleotides in length) and generate large quantities of elongated, theoretically single-stranded DNA or RNA molecules, which are periodically complementary to the template sequences. The synthesized DNA and RNA reach several kilobase pairs in molecular weight after a few hours of reaction, and they tend to form spherical particles by nucleic acid condensation resulting from inter- or intramolecular forces. Highlighted by the large amount of nucleic acid as well as the versatility of its technique, RCR has become one of the most promising approaches to construct nucleic acid-based nanostructures. The versatility of RCR has been demonstrated with the design of various functional DNA nanostructures including microsponges,14 nanoflowers,15,16 nanoclews,17,18 nanococoons,19 along with RNA nanostructures, such as microsponges,20,21 free-standing membranes,22 siRNA nanosheets,23 nanovectors,24 and RNA cargos.25 Therefore, this rapid and specific isothermal amplification reaction has led to the application of nucleic acid nanostructures as for example therapeutic and imaging agents.
In this work, by taking advantage of RCR, we developed robust and straightforward approaches to fabricate multifunctional organic–inorganic hybrid RNA particles, facilitating direct immobilization of various payloads through the selected chemistries. Hybrid RNA particles with a flower-like morphology (termed hRNFs hereafter) were synthesized by RCT, where complexation of inorganic Mg2PPi crystals and RNA strands resulted in the growth of RNA spheres in a time-dependent manner. The hRNFs comprised highly compact, multilayered thin petals that branch hierarchically outward from an inner core. Functional hRNFs were made by the in situ incorporation of modified uridine triphosphates (UTP) during the reaction or by electrostatic adsorption of positively charged proteins. This allowed selective surface decoration with various functional units such as small fluorescent dyes, proteins, quantum dots (QDs), and gold nanoparticles (GNPs). Moreover, the structural feature of as-synthesized hRNFs including large surface area, ease of surface functionalization, unique local environments created by the high charge density of RNA and abundant magnesium ions provided by Mg2PPi crystals, make them ideal platforms to immobilize biological molecules such as enzymes. To explore this, we coupled β-galactosidase (β-gal) enzymes to hRNFs and observed enhanced enzymatic activity and improved stability in comparison to free enzymes.
2. Results and Discussion
2.1. Controlled Synthesis of Porous Hybrid RNA Flowers (hRNFs)
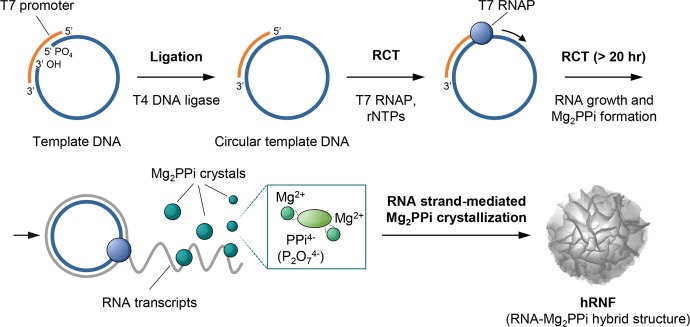
The construction of RNA structures was achieved via enzyme-assisted RCT amplification as illustrated in Figure 1. First, a 5′-phosphorylated linear template DNA was hybridized with a T7 promoter and further ligated with T4 DNA ligase to form a circular DNA. The resulting products were confirmed by both native and denatured polyacrylamide gel electrophoresis (Figure S1). Then, the as-synthesized circular DNA template was transcribed to synthetic RNA by a DNA-dependent RNA polymerase (RNAP), here T7 RNAP, in the presence of ribonucleotide triphosphates (rNTPs) in the reaction buffer at 37 °C, generating amplified single-stranded RNA with a high molecular weight. In this process, each ribonucleotide was covalently bound into the growing RNA chain, releasing pyrophosphate ions (PPi4– or P2O74–) simultaneously. Divalent metal cations, here magnesium ions (Mg2+), are crucial in polymerase-catalyzed nucleotidyl transfer reactions, both as activators for nucleophiles and as electrostatic stabilizers for negative charges of nucleic acids.26 The Mg2+ ions coordinate with phosphates, acidic residues of a polymerase, and water molecules, assisting the release of PPi, whereas free Mg2+ also easily binds to PPi4–, forming inorganic magnesium pyrophosphate (Mg2PPi or Mg2P2O7). Subsequently, the interactions between Mg2PPi and RNA lead to the formation of hybrid nanomaterials consisting of RNA and Mg2PPi, where long flexible RNA strands can actively mediate the nucleation and growth process of Mg2PPi precipitates.27,28 Consequently, RNA-based structures were formed predominantly through a nucleic-acid-driven Mg2PPi crystallization process with a minor contribution from Watson–Crick base pairing,15,27 leading to RNA–inorganic hybrid particles with characteristic structural properties. We further performed a series of optimization experiments to maximize the performance of enzymatic transcription, where the yield of RNA and PPi in the hRNF products was found to be affected by the concentration of reaction components, including the concentrations of rNTPs (Figure S2), template DNA (Figure S3), and T7 RNAP (Figure S4). Interestingly, the size of hRNFs could also be controlled by adjusting the concentration of the above components (Figures S2 and S3). To obtain relatively monodispersed hRNFs with sufficient amounts of RNA and Mg2PPi, we performed the optimal RCT reaction in a 50 μL solution containing circular template DNA (0.6 μM), rNTPs (2 mM), and T7 RNAP (5 U/μL) at 37 °C for 20 h.
Figure 1.
Schematic illustration of the proposed mechanism of RCT-mediated hRNF formation. The formation of hRNF particles results from the electrostatic attraction and van der Waals forces between RNA, magnesium (Mg2+) and pyrophosphate ions (PPi4– or P2O74–), in which long synthetic RNA strands actively participate in modulating the nucleation and growth process of Mg2PPi crystals.
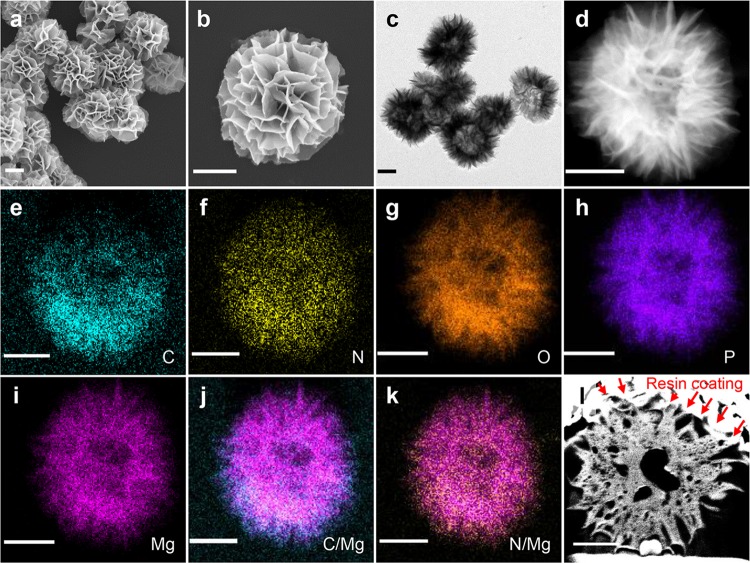
Structural characterization of RNA products was performed by scanning electron microscopy (SEM) and transmission electron microscopy (TEM) analysis. As shown in Figure 2a, b, the synthesized RNA products displayed a flower-like shape with petal structures (hereby termed as RNA flowers, RNFs), and were found to be uniformly 1–2 μm in diameter. The compositions and structures of the particles were further characterized by TEM imaging and elemental mapping using a scanning transmission electron microscopy (STEM) equipped with a high-angle annular dark-field (HAADF) detector (Figure 2c–k). The porous and hierarchical structure of the RNF is clearly observable in the HAADF-STEM image (Figure 2d). Energy-dispersive X-ray spectroscopy (EDS) mapping further showed that carbon (C), nitrogen (N), oxygen (O), magnesium (Mg), and phosphorus (P) were present in the particle, which confirmed the presence of RNA and Mg2PPi in hRNFs. By merging the elemental distribution of C and N with Mg, C and N were also found to be localized in the outer shell around the particle. These particles therefore represent hybrid materials consisting of organic (RNA) and inorganic (Mg2PPi) networks, hereby referred to as hybrid RNFs (hRNFs).
Figure 2.
Characterization of the hRNFs. Representative (a, b) SEM images, (c) TEM image, (d) HAADF-STEM image, (e–i) STEM-EDS elemental maps (C, N, O, P, and Mg) corresponding to d, merged images of (j) C and Mg maps, (k) N and Mg maps, and (l) SEM image of the orthogonal cross-section of a single hRNF sliced by FIB. Scale bars: 500 nm.
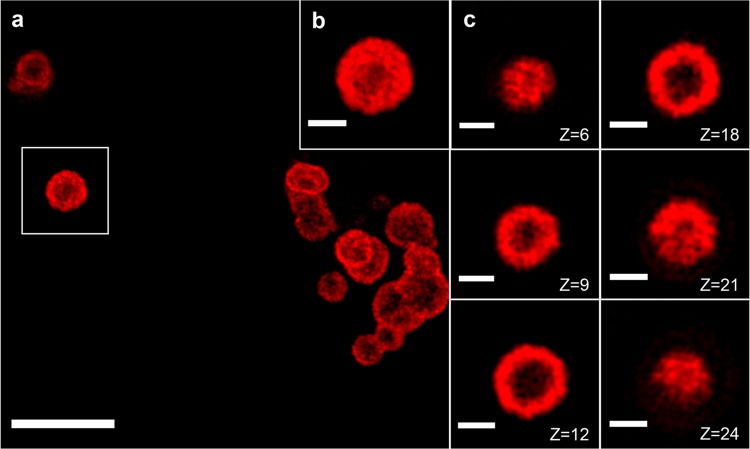
It is worth noting that apparent areas of bright and dark contrast inside the hRNFs were observed from the TEM and STEM images, respectively (Figure 2c, d), suggesting the existence of hollow cavities inside the hRNFs. To confirm this, we sectioned an hRNF particle by focused ion beam (FIB) and imaged it with SEM. This “slice and view” technique enables simultaneous electron microscopy imaging while the ion beam is milling the sample. With this method, the inner structure of hRNFs could be directly visualized once the cross-section was exposed. The hRNFs were embedded in resin to preserve the topological and morphological features of the sample, followed by sputter coating with chromium. With cross-sectioning by FIB and imaging by SEM, we observed that a central cavity with various nanopores appeared to be spread throughout the particle (Figure 2l). To further explore this structural feature, we carried out a transcription reaction in the presence of cyanine 5 dye (Cy5)-labeled UTP, allowing direct replacement of rUTP with Cy5-UTP and subsequent production of fluorescently labeled RNA. After purification, the localization of fluorescent RNA strands within the particles was examined by structured illumination microscopy (SIM).29,30 As shown in the z-stacks of the SIM images (Figure 3), the Cy5 fluorescence was predominantly localized on the surface of the particles, rather than homogeneously distributed within the particles. These results indicate that the prepared hRNFs have a characteristic internal cavity with a highly porous structure and present a high content of RNA strands on the periphery of the particle surface.
Figure 3.
Characterization of Cy5-labeled hRNFs by SIM imaging. (a) Representative SIM image of Cy5-labeled hRNFs and (b) a higher-magnification view of the particle in the white box in a. (c) Individual frames of the z-stack (step size: 100 nm) of the hRNF particle in b are shown in the right panel. Scale bars: 5 μm (main panel), 1 μm (inset and right panels).
2.2. Approaches toward hRNF Functionalization
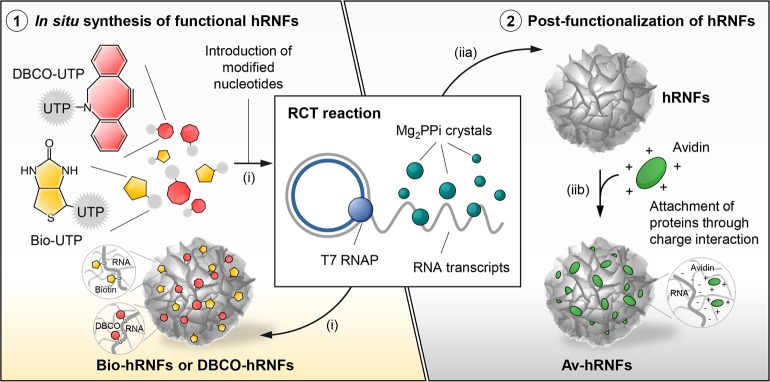
The fabrication of hRNFs through RCT enables the incorporation of versatile functional groups on the surface of the RNA structures. Herein, we demonstrated two strategies for hRNF functionalization, namely: covalently incorporating a chemical group (i.e., biotin and DBCO) using modified-UTP (i.e., biotin-UTP and DBCO-UTP) during the RCT reaction, and electrostatically assembling positive biomolecules (i.e., avidin) on hRNFs (Figure 4).
Figure 4.
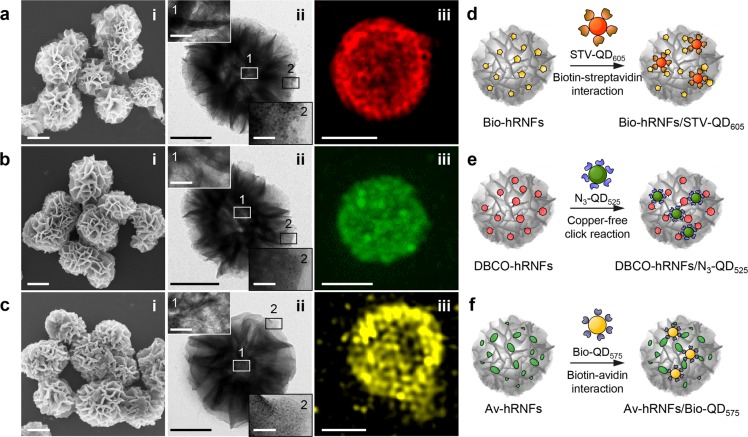
Schematic illustration of two synthetic approaches toward hRNF functionalization: (i) in situ introducing modified uridine triphosphates (UTP) (i.e., biotin-UTP and DBCO-UTP) during the RCT reaction and (iia, iib) electrostatically adsorbing positively charged proteins (i.e., avidin) onto the as-synthesized hRNFs to which biotinylated cargos can bind via biotin–avidin recognition. Biotin- or DBCO-conjugated UTP (Bio-UTP or DBCO-UTP); biotin-, DBCO-, or avidin-modified hRNFs (Bio-hRNFs, DBCO-hRNFs, or Av-hRNFs).
In a first approach, we introduced modified-UTP units (biotin- or DBCO-coupled UTP) into the RCT reaction mixtures, which were expected to be directly incorporated into the RNA sequences in place of the equivalent natural counterpart UTP. Consequently, the elongated RNA building blocks carried a sufficient amount of concatemers with functional moieties to offer numerous binding sites on the compact RNA particles for further modification. However, the relative RNA yield of the RCT reaction was found to decrease upon the addition of Bio-UTP or DBCO-UTP to the RCT reaction mixtures (Figure S5). This could be attributed to restrictions of steric mechanisms that RNA polymerase utilizes for ensuring accurate nucleotide incorporation and genome replication.31,32 Modifications on bases would easily increase the error rate of the T7 RNA polymerase and affect the reverse transcriptase fidelity.33 Therefore, to achieve both high production of RNA and functional groups within the functional hRNFs, we chose 40 μM as the optimal concentration for both UTPs in this work (Figure S5).
The incorporation of biotin and DBCO groups in the RCT did not cause notable structural changes or size variations in the hRNFs (Figure 5a, b). The presence of these two functional groups (biotin and DBCO) on the functionalized hRNFs was confirmed by the conjugation of QDs.34 As shown in Figure 5, streptavidin-conjugated QDs (STV-QD605) and azide-functionalized QDs (N3-QD525) could be coupled onto Bio-hRNFs (Figure 5a) and DBCO-hRNFs (Figure 5b) via biotin–streptavidin interaction and copper-free azide-DBCO click chemistry, respectively. The magnified TEM images of hRNF particles (site 1, core region; site 2, edge region) showed that QDs were predominantly located on the periphery of the particle surface, especially on the petals of the hRNFs (site 2). As further confirmed by SIM analysis (Figure 5a and Figure S6), fluorescence of QD605-labeled Bio-hRNFs appeared to distribute throughout the outer layer of the particle, which is very similar to the fluorescence signals from Cy5-labeled hRNFs (Figure 3). These results demonstrate that the Bio and DBCO-UTP groups were successfully incorporated in the RNA sequences of hRNFs, allowing selective interaction with STV-QD605 and N3-QD525.
Figure 5.
Surface functionalization of hRNFs: (a) biotin-labeled hRNFs (Bio-hRNFs), (b) DBCO-labeled hRNFs (DBCO-hRNFs), and (c) avidin-coated hRNFs (Av-hRNFs). (i) SEM images of functional hRNFs, and (ii) TEM images and (iii) SIM images of QD-labeled functional hRNFs. The insets in the TEM images represent the magnified views of the selected region denoted as 1 and 2 in each particle. (d–f) Schematic illustration showing labeling of each functional hRNFs with surface-modified QDs through selective interactions. The functional hRNFs are labeled with QDs as follows: streptavidin-modified QDs (STV-QD605) for Bio-hRNFs, azide-modifed QDs (N3-QD525) for DBCO-hRNFs, and biotin-modified (Bio-QD575) for Av-hRNFs. Scale bars: 500 nm (SEM and TEM images), 50 nm (TEM insets), and 1 μm (SIM images).
Because of the broad applicability of biotin–streptavidin affinity, it is possible to label Bio-hRNFs with different cargos such as avidin-modified Alexa Fluor 488 (Av-AF488) and streptavidin-modified horseradish peroxidase (STV-HRP). The loading of biotin on Bio-hRNFs via direct incorporation during RCT was confirmed by a commercial biotin quantification kit (Figure S7). The SIM images showed well-distributed green fluorescence, suggesting successful conjugation of the Av-AF488 onto the Bio-hRNFs (Figure S8). The high affinity (Kd ≈ 10–4 mol/L)35 of biotin–avidin interaction ensured that the kinetics of fluorophore adsorption was fast and efficient. Next, the assembly of STV-HRP on Bio-hRNFs was achieved using a similar method. As shown in Figure S9, the addition of STV-HRP onto Bio-hRNFs slightly altered the morphology and surface roughness. STEM-EDS analysis revealed the structural and elemental features of the Bio-hRNFs after HRP coating (Figure S10). The HAADF-STEM image of the STV-HRP coupled Bio-hRNFs (STV-HPR/Bio-hRNFs) shows a similar porous and hierarchical structure as to that observed in the Bio-hRNFs. EDS analysis of the STV-HRP/Bio-hRNFs showed that the average atomic ratio of C, N, and O elements relative to Mg increased in STV-HRP/Bio-hRNFs compared to those in the hRNFs, whereas the P/Mg ratio in both particles remained approximately consistent. This confirmed the presence of STV-HRP on the surface of Bio-hRNFs.
In a second approach, the positive biomolecule avidin (isoelectric point of ∼10.5) was bound to negatively charged hRNFs via electrostatic interaction, leading to the formation of avidin-coated hRNFs (Av-hRNFs). Compared to unmodified hRNFs, the surface roughness and the petal thickness of the Av-hRNFs were increased (Figure 5c and Figure S11). Significant morphological changes of the Av-hRNFs were observed upon varying the mass ratio of avidin to RNA in hRNFs. At the 1:1 ratio, the hRNF petals were found to be thicker, whereas the surface morphology was maintained (Figure S11b). A further increase in hRNF petal thickness was observed with the increase of the avidin to RNA ratio (2:1 and 5:1, Figure S11c, d). The porous features of the hRNFs were fully blocked by the adsorbed avidin molecules when the avidin to RNA ratio reached 10:1 (Figure S11e). The STEM-EDS analysis further supported the presence of both avidin and RNA on the Av-hRNFs. Compared to hRNFs (Figure 2d), Av-hRNFs with the dense coverage of avidin showed a less porous and hierarchical structure in HAADF-STEM images (Figure S12a). Merged EDS mapping of C and N with Mg yielded a further evidence for the avidin attachment. The average atomic ratios of C, N, O, and P normalized to Mg in Av-hRNFs were higher than those in hRNFs, which we attribute to the addition of avidin to the hRNFs, whereas the ratio of P to Mg remained unchanged (Figure S12b–i). The coating of avidin on hRNFs therefore enabled successful labeling of QDs (Figure 5c) and GNPs (Figure S13).
2.3. Kinetics of Enzymatic Reactions on Bio-hRNFs
Hybrid assemblies of nucleic acids and proteins have been constructed for fundamental understanding of nucleic acid–protein interactions as well as for use in various biomedical applications.2,36,37 Very interesting opportunities in the design of protein-nucleic acid complexes exist to develop efficient enzyme incorporation strategies, in which the enzymes could be encapsulated or attached via noncovalent or covalent bonding. Recently, new classes of organic–inorganic hybrid nanoflowers that integrate organic components, such as proteins and enzymes, and inorganic ions (i.e., Cu2+, Ca2+, and Fe2+) have been created by bioinspired mineralization methods.38−42 The enzymes incorporated into such types of hybrid materials have shown significantly improved enzyme activity and biological stability to free ones, offering their potential use in biocatalytic systems.
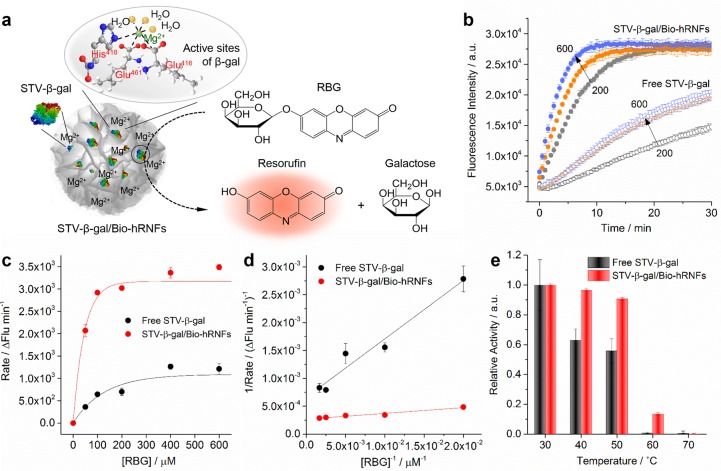
In this study, we focused on the remarkable features that hRNFs are not only highly adaptable structures for assembly with proteins but also comprise RNA and magnesium ions. Thus, the versatile functionalization and hydrophilic nature of the hRNFs make them an attractive platform for enzyme immobilization. Of note, the presence of surface-bound water, highly negative charges, and magnesium ions are anticipated to exert an important effect in regulating enzyme activity and stability. To explore this, the enzyme β-gal (streptavidin-conjugated β-gal used in this study, referred to as STV-β-gal hereafter) was selected to be immobilized on the Bio-hRNFs, and the enzymatic activity was studied. β-gal is an essential lysosomal enzyme involving in the breakdown of glycosphingolipids (e.g., GM1 ganglioside),43 and its deficiency can result in GM1 gangliosidosis, a lysosomal storage disorder.44 Moreover, as a typical allosteric enzyme, β-gal can be activated by magnesium ions, undergoing a conformation change upon metal ion binding. Nonfluorescent resorufin-β-d-galactopyranoside (RBG) was chosen as a model substrate, and the activity of β-gal can be monitored by the fluorescence emission at 584 nm because of the enzyme-catalyzed hydrolysis of RBG (nonfluorescent) into resorufin (red-fluorescent) and galactose (Figure 6a).
Figure 6.
(a) Schematic illustration of the STV-β-gal activity assay using the RBG substrate. The catalytic activity of STV-β-gal was measured by monitoring the red fluorescence signal of resorufin with an emission maximum at 584 nm. The active sites of β-gal containing a Mg2+ ion bound to three amino acid residues (His418, Glu416, and Glu461) and three water molecules are shown. (b) Catalytic kinetics of free STV-β-gal and STV-β-gal/Bio-hRNFs at different enzyme concentrations (200, 400, and 600 ng/mL). (c) Michaelis–Menten and (d) Lineweaver–Burk plots of free STV-β-gal and STV-β-gal/Bio-hRNFs. (e) Relative activity of free STV-β-gal and STV-β-gal/Bio-hRNFs at various temperatures (30–70 °C). The relative enzymatic activity was obtained by normalizing the fluorescence signals of both free STV-β-gal and STV-β-gal/Bio-hRNFs to that of free STV-β-gal or STV-β-gal/Bio-hRNFs heated at 30 °C, respectively. Results represent mean ± s.d. of three independent experiments.
To prepare β-gal enzyme-loaded hRNFs, we incubated STV-β-gal with Bio-hRNFs for 30 min prior to the addition of RBG substrate. The resulting STV-β-gal/Bio-hRNFs were directly used for kinetic experiments without further purification so as to keep the same enzyme concentration in both free STV-β-gal and STV-β-gal/Bio-hRNFs. β-gal exists as an active homotetrameric enzyme with four subunits and undergoes transitions from active tetramers to inactive monomers upon thermal denaturation.45 These conformational transitions of β-gal along with its activity are greatly affected by divalent cations such as Mg2+.46 We therefore hypothesized that efficient recruitment and surface immobilization of STV-β-gal on Mg2+-rich hRNF constructs may assist the enzyme activation and stabilization. To verify this, the kinetic experiments were conducted in magnesium-free PBS buffer (pH 7.4). As shown in Figure 6b, depending on the concentrations of STV-β-gal (200, 400, and 600 ng/mL), the fluorescence of the resorufin was observed to increase immediately after the addition of RBG substrate and reached a plateau after 20 min. A higher amount of enzyme generated fluorogenic enzymatic products at a faster rate. Interestingly, the free STV-β-gal displayed much lower catalytic activity against RBG hydrolysis as the fluorescence increase was obviously slow, and the reaction was not complete even after 30 min (Figure 6b). The Michaelis–Menten constant Km and the maximum reaction velocity Vmax of STV-β-gal were calculated using the Michaelis–Menten and Lineweaver–Burk plots (Figure 6c, d). As shown in Table 1, the STV-β-gal/Bio-hRNFs yielded a Km of 38.6 ± 5.7 μM, approximately 4.1-fold lower than that of free STV-β-gal (160.5 ± 31.5 μM). In addition, the Vmax of STV-β-gal/Bio-hRNFs was determined to be 3736.0 ± 77.1 ΔFlu/min, 2.5 times higher than that of free STV-β-gal (1522.6 ± 133.3 ΔFlu/min), suggesting that the immobilized STV-β-gal on the Bio-hRNFs had a higher efficiency in converting RBG into its products. The initial reaction velocities (v) of both free STV-β-gal and STV-β-gal/Bio-hRNFs were obtained from the kinetics curves in the presence of different RBG concentrations (Figure S14). Under the same test conditions with 400 μM of RBG, the overall activity of the immobilized STV-β-gal in the STV-β-gal/Bio-hRNFs was ca. 6.6–11.2 fold higher than that of free enzyme at the same enzyme concentration. Moreover, at a fixed enzyme concentration, the catalytic activity of β-gal was found to be dependent on the concentration of Bio-hRNFs, where a higher concentration of Bio-hRNFs resulted in a faster hydrolysis reaction (Figure S15). Finally, the thermostability of STV-β-gal was compared with that entrapped in Bio-hRNFs over the temperature range of 30–70 °C. As shown in Figure 6e, both the free STV-β-gal and STV-β-gal/Bio-hRNFs showed high activity after incubation at 30 °C. However, the enzymatic activity of free STV-β-gal dramatically dropped to ca. 60% after incubation at 40 °C, whereas the activity of STV-β-gal/Bio-hRNFs remained ca. 90% of the enzyme activity at 30 °C. Further, increases in temperature to (above 60 °C) resulted in little catalytic activity of the enzyme in both systems (Figure 6e).
Table 1. Kinetic Data Showing the Michaelis–Menten Constant (Km) and Maximum Velocity (Vmax) of Free STV-β-gal and STV-β-gal/Bio-hRNFsa.
| sample | Km (μM) | Vmax (ΔFlu/min) |
|---|---|---|
| free STV-β-gal | 160.5 ± 31.5 | 1522.6 ± 133.3 |
| STV-β-gal/Bio-hRNFs | 38.6 ± 5.7 | 3736.0 ± 77.1 |
Data represent mean ± s.d. of three independent experiments.
Clearly, the catalytic activity of the STV-β-gal immobilized on the Bio-hRNFs was substantially enhanced compared to free STV-β-gal. We ascribe this to the unique environment of the hRNFs, particularly the high charge density of RNA strands, large amount of surface-bound water, and abundant Mg2+ ions provided by either the Mg2PPi crystals or Mg2+-stabilized RNA molecules. First, enzymes immobilized on the hRNFs were extensively exposed to an environment full of negative charges that may resemble the relative abundance of polyanionic nucleic acid. As reported previously, the negative charges on large nucleic acid structures, such as in DNA origami, play an important role in regulating the activity of conjugated enzymes.36 Second, the hRNFs are expected to attract a strongly bound hydration layer, and in turn to form a hydrogen-bonded water molecule network around the hRNFs, because phosphate is a known kosmotropic anion that increases the extent of hydrogen-bonded water structures.2 Enzymes are likely to be more stable and active in a highly ordered hydrogen-bonded water environment. Third, it is well-known that cationic ions, such as monovalent or divalent ions, are of great importance in the regulation of enzymatic function by coordinating protein residues, functional groups of substrates, and water molecules. In particular, Mg2+ is known to play an important role in securing the active site of the β-gal, thereby protecting the active form of the enzyme from being thermally unfolded.47,48 The active sites of enzymes tend to be more susceptible to denaturation than their native forms as a whole, indicating that Mg2+-free or Mg2+-deficient β-gal could be easily unfolded by heating, rapidly losing its activity. It has been well documented that monovalent (i.e., Na+ and K+)49 and divalent (i.e., Mg2+)50 cations are essential to increase catalytic activity and binding affinity for substrates. Concerning the Mg2+-rich microenvironments around the hRNFs, the strong dependence of β-gal activity upon exposure to Mg2+ is an important consideration. Mg2+ has a strong influence on both the structure and function of β-gal. As previously reported, there are at least two Mg2+ binding sites within the β-gal enzyme. The Mg2+ that binds to the primary Mg2+ site (ligated by Glu416, His418, Glu461, and three water molecules, as shown in Figure 6a) can cause Km to decrease and modulate the chemistry of the active site.51,52 The metal binding would also cause a conformational change of the enzyme, which is believed to benefit its structural stability.53 Because of the positive modulation effects of Mg2+, the immobilized STV-β-gals stayed in an active and stable form.
3. Conclusion
In summary, we have presented enzyme-assisted, RCT-driven RNA assembly into flower-shaped porous structures, termed as hRNFs, and demonstrated their use as a loading platform for biomolecules. We systematically investigated the composition and structure of hRNFs with combined characterization techniques including (S)TEM, FIB-SEM, and SIM and showed that the hierarchical porous structures were formed as a hybrid composite of organic and inorganic RNA/Mg2PPi species. The RCT approach provided significant advantages for surface functionalization by in situ incorporation of functional groups (i.e., DBCO and biotin) into the hRNFs, allowing surface modification via copper-free click chemistry and biotin–avidin affinity. We also demonstrated a postfunctionalization method for effective surface conjugation through electrostatic interactions and biotin–avidin recognition. Because of the high surface area, the hRNFs offered a large capacity for on-demand payload immobilization. Furthermore, we showed that hRNFs increased the catalytic activity and thermal stability of STV-β-gal enzymes immobilized onto the surface, which was ascribed to the unique local environment created by the high charge density of RNA strands and abundant Mg2+ ions. We envision that these RNA-based hybrid structures can be further engineered as versatile platforms for a broad range of biological targets such as small molecules, nucleic acids, and proteins, which will lead to potential applications in biomedicine. For example, the hRNFs can be developed into a delivery system for intracellular delivery of β-gal, which is considered as one of the preferable methods to treat lysosomal storage disorder.
Acknowledgments
Y.W. acknowledges support from the Imperial-CSC Scholarship. E.K. acknowledges support from Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education (2015R1A6A3A03018919). W.K.-A. was supported by the British Heart Foundation Centre of Research Excellence (RE/13/4/30184). S.G was funded by a PhD studentship in Biomedicine and Bioengineering in Osteoarthritis, Imperial College London. P.D.H. acknowledges support from European Union’s Horizon 2020 research and innovation programme through the Individual Marie Skłodowska-Curie Fellowship “Ampidots” (701994). M.M.S. acknowledges the ERC Seventh Framework Programme Consolidator grant “Naturale CG” (616417) and the grant “Bio-functionalized Nanomaterials for Ultra-sensitive Biosensing” (EP/K020641/1) funded by Engineering and Physical Science Research Council (EPSRC). We acknowledge Vincent Leonardo for his great suggestions on SIM imaging. The authors acknowledge use of the characterization facilities within the Harvey Flower Electron Microscopy Suite (Department of Materials) at Imperial College London, and the Facility for Imaging by Light Microscopy at Imperial College London, which is partially funded by the Biotechnology and Biological Science Research Council (BBSRC, BB/L015129/1).
Supporting Information Available
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acsami.9b04663.
Details of materials and methods, gel electrophoresis analysis, optimization studies of the RCT reaction, SIM images of STV-QD605/Bio-hRNFs and Av-AF488/Bio-hRNFs, quantification of biotin concentration in Bio-hRNFs, characterization of STV-HRP/Bio-hRNFs and Av-hRNFs by SEM/STEM imaging and STEM-EDS analysis, SEM and TEM images of Bio-GNPs/Av-hRNFs, kinetics study of STV-β-gal and STV-β-gal/Bio-hRNFs complexes (PDF)
Author Present Address
‡ P.D.H. is currently at Department of Chemistry and Applied Biosciences, ETH Zürich, CH-8093 Zürich, Switzerland.
Author Contributions
† Y.W. and E.K. contributed equally.
The authors declare no competing financial interest.
Notes
Research data is available at DOI: 10.5281/zenodo.3243963.
Supplementary Material
References
- Smith D.; Schüller V.; Engst C.; Rädler J.; Liedl T. Nucleic Acid Nanostructures for Biomedical Applications. Nanomedicine 2013, 8, 105–121. 10.2217/nnm.12.184. [DOI] [PubMed] [Google Scholar]
- Zhao Z.; Fu J.; Dhakal S.; Johnson-Buck A.; Liu M.; Zhang T.; Woodbury N. W.; Liu Y.; Walter N. G.; Yan H. Nanocaged Enzymes with Enhanced Catalytic Activity and Increased Stability Against Protease Digestion. Nat. Commun. 2016, 7, 10619. 10.1038/ncomms10619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Y.; Roberts C. C.; Zhu J.; Lin J.-L.; Chang C.-e. A.; Wheeldon I. Tuning Enzyme Kinetics Through Designed Intermolecular Interactions Far From the Active Site. ACS Catal. 2015, 5, 2149–2153. 10.1021/acscatal.5b00130. [DOI] [Google Scholar]
- Rothemund P. W. K. Folding DNA to Create Nanoscale Shapes and Patterns. Nature 2006, 440, 297–302. 10.1038/nature04586. [DOI] [PubMed] [Google Scholar]
- Hong F.; Zhang F.; Liu Y.; Yan H. DNA Origami: Scaffolds for Creating Higher Order Structures. Chem. Rev. 2017, 117, 12584–12640. 10.1021/acs.chemrev.6b00825. [DOI] [PubMed] [Google Scholar]
- Fu J.; Liu M.; Liu Y.; Woodbury N. W.; Yan H. Interenzyme Substrate Diffusion for an Enzyme Cascade Organized on Spatially Addressable DNA Nanostructures. J. Am. Chem. Soc. 2012, 134, 5516–5519. 10.1021/ja300897h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilner O. I.; Weizmann Y.; Gill R.; Lioubashevski O.; Freeman R.; Willner I. Enzyme Cascades Activated on Topologically Programmed DNA Scaffolds. Nat. Nanotechnol. 2009, 4, 249–254. 10.1038/nnano.2009.50. [DOI] [PubMed] [Google Scholar]
- Fu J.; Yang Y. R.; Johnson-Buck A.; Liu M.; Liu Y.; Walter N. G.; Woodbury N. W.; Yan H. Multi-Enzyme Complexes on DNA Scaffolds Capable of Substrate Channelling with an Artificial Swinging Arm. Nat. Nanotechnol. 2014, 9, 531–536. 10.1038/nnano.2014.100. [DOI] [PubMed] [Google Scholar]
- Linko V.; Nummelin S.; Aarnos L.; Tapio K.; Toppari J. J.; Kostiainen M. A. DNA-Based Enzyme Reactors and Systems. Nanomaterials 2016, 6, 139. 10.3390/nano6080139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romashchenko A. V.; Kan T.-W.; Petrovski D. V.; Gerlinskaya L. A.; Moshkin M. P.; Moshkin Y. M. Nanoparticles Associate with Intrinsically Disordered RNA-Binding Proteins. ACS Nano 2017, 11, 1328–1339. 10.1021/acsnano.6b05992. [DOI] [PubMed] [Google Scholar]
- Guo P. The Emerging Field of RNA Nanotechnology. Nat. Nanotechnol. 2010, 5, 833–842. 10.1038/nnano.2010.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y.; Chen F.; Li Q.; Wang L.; Fan C. Isothermal Amplification of Nucleic Acids. Chem. Rev. 2015, 115, 12491–12545. 10.1021/acs.chemrev.5b00428. [DOI] [PubMed] [Google Scholar]
- Mohsen M. G.; Kool E. T. The Discovery of Rolling Circle Amplification and Rolling Circle Transcription. Acc. Chem. Res. 2016, 49, 2540–2550. 10.1021/acs.accounts.6b00417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roh Y. H.; Lee J. B.; Shopsowitz K. E.; Dreaden E. C.; Morton S. W.; Poon Z.; Hong J.; Yamin I.; Bonner D. K.; Hammond P. T. Layer-by-Layer Assembled Antisense DNA Microsponge Particles for Efficient Delivery of Cancer Therapeutics. ACS Nano 2014, 8, 9767–9780. 10.1021/nn502596b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu G.; Hu R.; Zhao Z.; Chen Z.; Zhang X.; Tan W. Noncanonical Self-Assembly of Multifunctional DNA Nanoflowers for Biomedical Applications. J. Am. Chem. Soc. 2013, 135, 16438–16445. 10.1021/ja406115e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lv Y.; Hu R.; Zhu G.; Zhang X.; Mei L.; Liu Q.; Qiu L.; Wu C.; Tan W. Preparation and Biomedical Applications of Programmable and Multifunctional DNA Nanoflowers. Nat. Protoc. 2015, 10, 1508–1524. 10.1038/nprot.2015.078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun W.; Jiang T.; Lu Y.; Reiff M.; Mo R.; Gu Z. Cocoon-Like Self-Degradable DNA Nanoclew for Anticancer Drug Delivery. J. Am. Chem. Soc. 2014, 136, 14722–14725. 10.1021/ja5088024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun W.; Ji W.; Hall J. M.; Hu Q.; Wang C.; Beisel C. L.; Gu Z. Self-Assembled DNA Nanoclews for the Efficient Delivery of CRISPR–Cas9 for Genome Editing. Angew. Chem., Int. Ed. 2015, 54, 12029–12033. 10.1002/anie.201506030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C.; Sun W.; Wright G.; Wang A. Z.; Gu Z. Inflammation-Triggered Cancer Immunotherapy by Programmed Delivery of CpG and Anti-PD1 Antibody. Adv. Mater. 2016, 28, 8912–8920. 10.1002/adma.201506312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J. B.; Hong J.; Bonner D. K.; Poon Z.; Hammond P. T. Self-Assembled RNA Interference Microsponges for Efficient siRNA Delivery. Nat. Mater. 2012, 11, 316–322. 10.1038/nmat3253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roh Y. H.; Deng J. Z.; Dreaden E. C.; Park J. H.; Yun D. S.; Shopsowitz K. E.; Hammond P. T. A Multi-RNAi Microsponge Platform for Simultaneous Controlled Delivery of Multiple Small Interfering RNAs. Angew. Chem., Int. Ed. 2016, 55, 3347–3351. 10.1002/anie.201508978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han D.; Park Y.; Kim H.; Lee J. B. Self-assembly of Free-standing RNA Membranes. Nat. Commun. 2014, 5, 4367. 10.1038/ncomms5367. [DOI] [PubMed] [Google Scholar]
- Kim H.; Lee J. S.; Lee J. B. Generation of siRNA Nanosheets for Efficient RNA Interference. Sci. Rep. 2016, 6, 25146. 10.1038/srep25146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park Y.; Kim H.; Lee J. B. Self-assembled DNA-Guided RNA Nanovector via Step-Wise Dual Enzyme Polymerization (SDEP) for Carrier-Free siRNA Delivery. ACS Biomater. Sci. Eng. 2016, 2, 616–624. 10.1021/acsbiomaterials.5b00554. [DOI] [PubMed] [Google Scholar]
- Kim H.; Jeong J.; Kim D.; Kwak G.; Kim S. H.; Lee J. B. Bubbled RNA-Based Cargo for Boosting RNA Interference. Adv. Sci. 2017, 4, 1600523. 10.1002/advs.201600523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castro C.; Smidansky E. D.; Arnold J. J.; Maksimchuk K. R.; Moustafa I.; Uchida A.; Götte M.; Konigsberg W.; Cameron C. E. Nucleic Acid Polymerases Use a General Acid for Nucleotidyl Transfer. Nat. Struct. Mol. Biol. 2009, 16, 212–218. 10.1038/nsmb.1540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shopsowitz K. E.; Roh Y. H.; Deng Z. J.; Morton S. W.; Hammond P. T. RNAi-Microsponges Form Through Self-Assembly of the Organic and Inorganic Products of Transcription. Small 2014, 10, 1623–1633. 10.1002/smll.201302676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim E.; Agarwal S.; Kim N.; Hage F. S.; Leonardo V.; Gelmi A.; Stevens M. M. Bioinspired Fabrication of DNA–Inorganic Hybrid Composites Using Synthetic DNA. ACS Nano 2019, 13, 2888–2900. 10.1021/acsnano.8b06492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang B.; Babcock H.; Zhuang X. Breaking the Diffraction Barrier: Super-Resolution Imaging of Cells. Cell 2010, 143, 1047–1058. 10.1016/j.cell.2010.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim E.; Zwi-Dantsis L.; Reznikov N.; Hansel C. S.; Agarwal S.; Stevens M. M. One-Pot Synthesis of Multiple Protein-Encapsulated DNA Flowers and Their Application in Intracellular Protein Delivery. Adv. Mater. 2017, 29, 1701086. 10.1002/adma.201701086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson S. J.; Beese L. S. Structures of Mismatch Replication Errors Observed in a DNA Polymerase. Cell 2004, 116, 803–816. 10.1016/S0092-8674(04)00252-1. [DOI] [PubMed] [Google Scholar]
- Lee I.; Berdis A. J. Non-Natural Nucleotides as Probes for the Mechanism and Fidelity of DNA Polymerases. Biochim. Biophys. Acta, Proteins Proteomics 2010, 1804, 1064–1080. 10.1016/j.bbapap.2009.08.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potapov V.; Fu X.; Dai N.; Corrêa J. I. R.; Tanner N. A.; Ong J. L. Base Modifications Affecting RNA Polymerase and Reverse Transcriptase Fidelity. Nucleic Acids Res. 2018, 46, 5753–5763. 10.1093/nar/gky341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y.; Howes P. D.; Kim E.; Spicer C. D.; Thomas M. R.; Lin Y.; Crowder S. W.; Pence I. J.; Stevens M. M. Duplex-Specific Nuclease-Amplified Detection of MicroRNA Using Compact Quantum Dot–DNA Conjugates. ACS Appl. Mater. Interfaces 2018, 10, 28290–28300. 10.1021/acsami.8b07250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green N. M. Avidin. Adv. Protein Chem. 1975, 29, 85–133. 10.1016/S0065-3233(08)60411-8. [DOI] [PubMed] [Google Scholar]
- Zhang Y.; Tsitkov S.; Hess H. Proximity Does Not Contribute to Activity Enhancement in the Glucose Oxidase–Horseradish Peroxidase Cascade. Nat. Commun. 2016, 7, 13982. 10.1038/ncomms13982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins J.; Zhang T.; Oh S. W.; Maloney R.; Fu J. DNA-Crowded Enzyme Complexes with Enhanced Activities and Stabilities. Chem. Commun. 2017, 53, 13059–13062. 10.1039/C7CC07361E. [DOI] [PubMed] [Google Scholar]
- Ge J.; Lei J.; Zare R. N. Protein–Inorganic Hybrid Nanoflowers. Nat. Nanotechnol. 2012, 7, 428–432. 10.1038/nnano.2012.80. [DOI] [PubMed] [Google Scholar]
- Somturk B.; Hancer M.; Ocsoy I.; Özdemir N. Synthesis of Copper Ion Incorporated Horseradish Peroxidase-Based Hybrid Nanoflowers for Enhanced Catalytic Activity and Stability. Dalton Trans 2015, 44, 13845–13852. 10.1039/C5DT01250C. [DOI] [PubMed] [Google Scholar]
- Celik C.; Tasdemir D.; Demirbas A.; Katı A.; Gul O. T.; Cimen B.; Ocsoy I. Formation of Functional Nanobiocatalysts with a Novel and Encouraging Immobilization Approach and Their Versatile Bioanalytical Applications. RSC Adv. 2018, 8, 25298–25303. 10.1039/C8RA03250E. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ocsoy I.; Dogru E.; Usta S. A New Generation of Flowerlike Horseradish Peroxides as a Nanobiocatalyst for Superior Enzymatic Activity. Enzyme Microb. Technol. 2015, 75–76, 25–29. 10.1016/j.enzmictec.2015.04.010. [DOI] [PubMed] [Google Scholar]
- Wang L.-B.; Wang Y.-C.; He R.; Zhuang A.; Wang X.; Zeng J.; Hou J. G. A New Nanobiocatalytic System Based on Allosteric Effect with Dramatically Enhanced Enzymatic Performance. J. Am. Chem. Soc. 2013, 135, 1272–1275. 10.1021/ja3120136. [DOI] [PubMed] [Google Scholar]
- Juers D. H.; Jacobson R. H.; Wigley D.; Zhang X.-J.; Huber R. E.; Tronrud D. E.; Matthews B. W. High Resolution Refinement of β-Galactosidase in a New Crystal Form Reveals Multiple Metal-Binding Sites and Provides a Structural Basis for α-Complementatio. Protein Sci. 2000, 9, 1685–1699. 10.1110/ps.9.9.1685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandhoff K.; Harzer K. Gangliosides and Gangliosidoses: Principles of Molecular and Metabolic Pathogenesis. J. Neurosci. 2013, 33, 10195–10208. 10.1523/JNEUROSCI.0822-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards R. A.; Jacobson A. L.; Huber R. E. Thermal Denaturation of Beta-Galactosidase and of Two Site Specific Mutants. Biochemistry 1990, 29, 11001–11008. 10.1021/bi00501a019. [DOI] [PubMed] [Google Scholar]
- Hei D. J.; Clark D. S. Estimation of Melting Curves from Enzymatic Activity–Temperature Profiles. Biotechnol. Bioeng. 1993, 42, 1245–1251. 10.1002/bit.260421015. [DOI] [PubMed] [Google Scholar]
- Gallagher C. N.; Huber R. E. Monomer–Dimer Equilibrium of Uncomplemented M15 β-Galactosidase from Escherichia Coli. Biochemistry 1997, 36, 1281–1286. 10.1021/bi961347a. [DOI] [PubMed] [Google Scholar]
- Rickenberg H. V. The Effect of Metal Ions and Proteins on the Stability of the β-Galactosidase of Escherichia Coli. Biochim. Biophys. Acta 1959, 35, 122–129. 10.1016/0006-3002(59)90341-5. [DOI] [Google Scholar]
- Xu J.; McRae M. A. A.; Harron S.; Rob B.; Huber R. E. A Study of the Relationships of Interactions Between Asp-201, Na+ or K+, and Galactosyl C6 Hydroxyl and Their Effects on Binding and Reactivity of β-Galactosidase. Biochem. Cell Biol. 2004, 82, 275–284. 10.1139/o04-004. [DOI] [PubMed] [Google Scholar]
- Case G. S.; Sinnott M. L.; Tenu J. P. The Role of Magnesium Ions in Beta-Galactosidase Hydrolyses. Studies on Charge and Shape of the Beta-Galactopyranosyl Binding Site. Biochem. J. 1973, 133, 99–104. 10.1042/bj1330099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth N. J.; Huber R. E. GLU-416 of β-Galactosidase (Escherichia coli) Is a Mg2+ Ligand and β-Galactosidases with Substitutions for GLU-416 Are Inactivated, Rather than Activated, by Mg2+. Biochem. Biophys. Res. Commun. 1996, 219, 111–115. 10.1006/bbrc.1996.0190. [DOI] [PubMed] [Google Scholar]
- Juers D. H.; Matthews B. W.; Huber R. E. LacZ β-Galactosidase: Structure and Function of an Enzyme of Historical and Molecular Biological Importance. Protein Sci. 2012, 21, 1792–1807. 10.1002/pro.2165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tenu J.-P.; Viratelle O. M.; Yon J. Kinetic Study of the Activation Process of β-Galactosidase from Escherichia coli by Mg2+. Eur. J. Biochem. 1972, 26, 112–118. 10.1111/j.1432-1033.1972.tb01746.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.