Site-specific X-ray triggered photo-oxidation combined with temperature-controlled crystallography can provide a powerful means to reveal structural mechanisms of macromolecular function for cysteine-dependent enzymes.
Keywords: cysteine modification, radiation-controlled photo-oxidation, serial X-ray crystallography, enzyme conformational dynamics, molecular dynamics simulations
Abstract
Cysteine is a rare but functionally important amino acid that is often subject to covalent modification. Cysteine oxidation plays an important role in many human disease processes, and basal levels of cysteine oxidation are required for proper cellular function. Because reactive cysteine residues are typically ionized to the thiolate anion (Cys-S−), their formation of a covalent bond alters the electrostatic and steric environment of the active site. X-ray-induced photo-oxidation to sulfenic acids (Cys-SOH) can recapitulate some aspects of the changes that occur under physiological conditions. Here we propose how site-specific cysteine photo-oxidation can be used to interrogate ensuing changes in protein structure and dynamics at atomic resolution. Although this powerful approach can connect cysteine covalent modification to global protein conformational changes and function, careful biochemical validation must accompany all such studies to exclude misleading artifacts. New types of X-ray crystallography experiments and powerful computational methods are creating new opportunities to connect conformational dynamics to catalysis for the large class of systems that use covalently modified cysteine residues for catalysis or regulation.
1. Cysteine modifications are functionally important and can be driven by X-ray radiation
Most functionally important cysteine residues in proteins are covalently modified at some point during their cellular lives. Whether serving as a catalytic nucleophile or as a site for post-translational modification, the proclivity of cysteine for forming bonds has been capitalized upon by evolution to serve functional ends (Reddie & Carroll, 2008 ▸). Cysteine oxidative post-translational modification states range from sulfenate (–SO−), sulfinate (–SO2 −) and sulfonate (–SO3 −) (Reddie & Carroll, 2008 ▸). Because cysteine is most reactive in its ionized thiolate form (Cys-S−), such modifications necessarily alter the active site electrostatic environment. The unperturbed pK a of the cysteine thiol sidechain is 8.3. However, the protein structural environment of a reactive cysteine residue can reduce its pK a value to well below 7, suggesting reactive thiolates are the dominant species at physiological pH. Even in cases where a functionally important cysteine is not subject to direct modification, such as in metal binding sites, the electrostatic properties of the thiolate anion are critically important. Thiolates are good hydrogen bond acceptors, and the hydrogen bonding interactions of the cysteine Sγ atom largely determines its reactivity and functional role (Roos et al., 2013 ▸; Mazmanian et al., 2016 ▸). Surprisingly, how cysteine modification alters protein conformational ensembles and dynamics is largely unexplored.
High-intensity X-ray irradiation produces a reducing, electron-rich environment in macromolecular crystals (Garman & Weik, 2017 ▸). Accordingly, most commonly observed forms of radiation damage are reductive, including metal reduction and cleavage of disulfides. Nevertheless, one of the primary species created by X-ray irradiation of aqueous samples is the hydroxyl radical (HO ) (George et al., 2012 ▸), the most potently oxidizing reactive oxygen species (ROS). At room temperature in water, HO
) (George et al., 2012 ▸), the most potently oxidizing reactive oxygen species (ROS). At room temperature in water, HO has a diffusion-limited lifetime of ∼1 ns, and thus reacts with the first moiety it encounters (Winterbourn, 2008 ▸). Of the 20 canonical proteinogenic amino acids, cysteine is most sensitive to oxidative modification. X-ray induced photo-chemistry at a nearby water molecule can form a hydroxyl radical, which can further react to form hydrogen peroxide (H2O2) (George et al., 2012 ▸). The oxidation of Cys-S− to Cys-SOH, primarily using H2O2 as the oxidant in vivo, forms the basis for many intracellular redox signaling pathways (García-Santamarina et al., 2014 ▸). Importantly, not all cysteine residues are equally susceptible to modification by ROS. Instead, the protein environment is finely tuned to determine which modified cysteine species are favored and determines the rate of their modification (Marinho et al., 2014 ▸). Although X-ray-induced cysteine photo-oxidation is distinct from cysteine oxidation in vivo, there is growing evidence that the physico-chemical contributors to site-specific cysteine X-ray photooxidation overlap with physiological processes (e.g. Fig. 1 ▸) (Maleknia et al., 1999 ▸; Xu & Chance, 2005 ▸; Young et al., 2019 ▸; Wilson et al., 2003 ▸). While there are comparatively few validated examples of cysteine X-ray photooxidation in protein crystal structures, it may be more common than previously acknowledged. Residues other than cysteine are likely subject to photo-oxidation as well, and these modifications may be under-reported (Wang, 2016 ▸).
has a diffusion-limited lifetime of ∼1 ns, and thus reacts with the first moiety it encounters (Winterbourn, 2008 ▸). Of the 20 canonical proteinogenic amino acids, cysteine is most sensitive to oxidative modification. X-ray induced photo-chemistry at a nearby water molecule can form a hydroxyl radical, which can further react to form hydrogen peroxide (H2O2) (George et al., 2012 ▸). The oxidation of Cys-S− to Cys-SOH, primarily using H2O2 as the oxidant in vivo, forms the basis for many intracellular redox signaling pathways (García-Santamarina et al., 2014 ▸). Importantly, not all cysteine residues are equally susceptible to modification by ROS. Instead, the protein environment is finely tuned to determine which modified cysteine species are favored and determines the rate of their modification (Marinho et al., 2014 ▸). Although X-ray-induced cysteine photo-oxidation is distinct from cysteine oxidation in vivo, there is growing evidence that the physico-chemical contributors to site-specific cysteine X-ray photooxidation overlap with physiological processes (e.g. Fig. 1 ▸) (Maleknia et al., 1999 ▸; Xu & Chance, 2005 ▸; Young et al., 2019 ▸; Wilson et al., 2003 ▸). While there are comparatively few validated examples of cysteine X-ray photooxidation in protein crystal structures, it may be more common than previously acknowledged. Residues other than cysteine are likely subject to photo-oxidation as well, and these modifications may be under-reported (Wang, 2016 ▸).
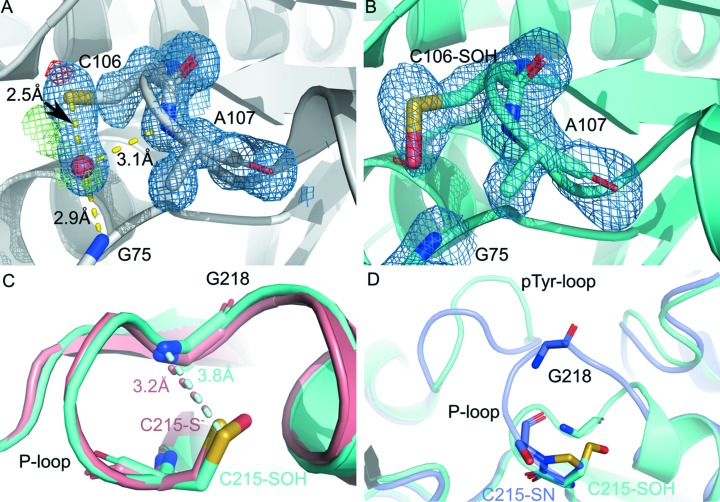
Figure 1.
Cysteine modification in health and disease. (A) Site-specific radiation damage at DJ-1 C106 (PDB ID 2or3, resolution 1.2 Å). A reactive cysteine near a water is a likely prerequisite for hydroxyl radical-mediated, site-specific photo-oxidation of Cys to Cys-SOH. Because of the hydroxyl radical’s extreme reactivity, it will modify the first moiety it contacts, for example a reactive cysteine. A water near C106, stabilized by amides of G75 and A107, is often observed in DJ-1 crystal structures. Difference density, contoured at 3σ, and 2F o − F c density, contoured at 1σ, that is elongated suggests partial oxidation of C106. A water molecule is represented as a red sphere. (B) Cys106-SOH is more fully formed. (PDB ID 3sf8; resolution 1.56 Å). (C) The catalytic cysteine C215 of PTP-1B is located in the P-loop, forming an S-NH hydrogen bond with G218 (salmon, PDB ID 2hnp, resolution 2.8 Å). Oxidation to C215-SOH neutralizes the negative charge of the C215 Sγ thiolate, making it a poorer hydrogen-bond acceptor and lengthening the hydrogen bond by nearly 20% (cyan, PDB ID 1oet, resolution 2.3 Å). (D) Further oxidation to C215-SN disrupts the C215-G218 hydrogen bond, concomitant with large conformational changes propagating to the nearby pTyr recognition loop (slate, PDB ID 1oes, resolution 2.2 Å; the C215-SOH conformation from panel A in cyan). All structures shown in this figure represent data collected at 100 K.
2. Cysteine oxidation in health and disease
Basal levels of cysteine oxidation are required for proper cellular function. Many proteins are modified by ROS to facilitate redox signaling, wound healing, growth and development. For example, recent data suggest that epidermal growth factor receptor (EGFR) is regulated by the reversible formation of a Cys-sulfenic acid (Truong et al., 2016 ▸), and glyceraldehyde 3-phosphate dehydrogenase (GAPDH) is a classic example of an enzyme where cysteine modification controls metabolic flux (Peralta et al., 2015 ▸). Numerous potent kinase inhibitors covalently modify conserved non-catalytic cysteine residues in the ATP binding sites (Backus, 2019 ▸). Indeed, the reactivity of accessible cysteines can be exploited for rational design of therapeutics (Bauer, 2015 ▸; Awoonor-Williams & Rowley, 2018 ▸; Gehringer & Laufer, 2019 ▸). Targeted covalent inhibitors (TCIs), which are small molecules that include a nucleophilic group to irreversibly bind to nucleophiles, are enjoying renewed interest. TCIs combine high affinity with selectivity (Casimiro-Garcia et al., 2018 ▸), and inhibit their target indefinitely. Potentially half of the human kinome could be targeted with TCIs (Liu et al., 2013 ▸), and covalent targeting of oncogenic missense mutations that introduce cysteine has been suggested as a promising avenue for personalized therapy (Ostrem et al., 2013 ▸; Visscher et al., 2016 ▸). Thus, identifying accessible, reactive cysteine residues and understanding how their covalent modification modulates protein conformational dynamics and function can impact rational drug design.
Human DJ-1 is an important therapeutic target that is regulated by cysteine oxidation. Impairment or absence of DJ-1 (PARK7) causes heritable parkinsonism (Bonifati et al., 2003 ▸) and its overexpression is correlated with invasive cancers (Nagakubo et al., 1997 ▸; Cao et al., 2015 ▸). DJ-1 dysregulation is also implicated in ischemia-reperfusion injury (Aleyasin et al., 2007 ▸) and diseases of the eye (Bonilha et al., 2017 ▸). Over a decade of work by many labs supports a role for DJ-1 in suppressing the endogenous formation of mitochondrially derived ROS (Guzman et al., 2010 ▸), defending against mitochondrial complex I inhibition (Canet-Avilés et al., 2004 ▸; Mullett & Hinkle, 2011 ▸), and activating pro-survival cellular responses (Aron et al., 2010 ▸). The conserved Cys106 residue is essential for these diverse functions of DJ-1 and has recently been targeted by two distinct classes of potent covalent inhibitors (Tashiro et al., 2018 ▸; Drechsel et al., 2018 ▸).
Oxidation of Cys106 allows DJ-1 to detect elevations in cellular ROS. Cys106 is also the nucleophile in the weak glyoxalase/deglycase activities of DJ-1 (Lee et al., 2012 ▸; Richarme et al., 2015 ▸) and may serve other catalytic roles as well. Cys106 has a low pK
a of 5.4 (Witt et al., 2008 ▸) and was first identified as a possible redox target owing to its high sensitivity to synchrotron radiation damage (Wilson et al., 2003 ▸). That study illustrated how site-specific radiation damage can serve as a guide for identifying reactive, functionally important cysteine residues in proteins. In fact, nearly all DJ-1 crystal structures determined to date show signs of site-specific radiation damage at Cys106 [e.g., Fig. 1(A) ▸], varying from ambiguous positive difference electron density to clearly resolved modifications such as Cys-SO2
− (Barbieri et al., 2018 ▸; Xu et al., 2018 ▸; Saito et al., 2014 ▸). Subsequent work showed that Cys106 readily oxidizes to Cys106-SO2
−
in vitro and in vivo (Canet-Avilés et al., 2004 ▸; Cao et al., 2014 ▸) which stabilizes DJ-1 and is important for preservation of mitochondrial morphology and resistance to complex I inhibition (Canet-Avilés et al., 2004 ▸; Witt et al., 2008 ▸; Cao et al., 2014 ▸; Joselin et al., 2012 ▸). In addition to the physiologically important formation of Cys106-SO2
−, partial oxidation of Cys106 to Cys106-SOH was observed in several X-ray crystallographic studies of DJ-1, which may be X-ray-driven modifications (Witt et al., 2008 ▸; Blackinton et al., 2009 ▸; Tashiro et al., 2014 ▸) [Fig. 1(B) ▸]. Cys106 is typically the only one of the three cysteine residues in DJ-1 that is oxidized upon X-ray illumination and is modified even in cryocooled crystals. This suggests that the probable HO oxidant is generated near Cys106, as HO
oxidant is generated near Cys106, as HO could not diffuse far owing to the vitrified buffer of a cryocooled crystal (Owen et al., 2012 ▸) and because it would react with the first residue it encountered.
could not diffuse far owing to the vitrified buffer of a cryocooled crystal (Owen et al., 2012 ▸) and because it would react with the first residue it encountered.
Another example of a protein where cysteine modification is functionally important is human tyrosine phosphatase 1B (PTP-1B), a major drug target for diabetes (Yip et al., 2010 ▸), cancer (Julien et al., 2011 ▸) and other diseases. PTP-1B is regulated by the reversible oxidation of its active site cysteine residue (C215) to a sulfenic acid (Salmeen et al., 2003 ▸; Böhmer et al., 2013 ▸). C215 is located within the P-loop, where its low pK a value of 5.4 (Lohse et al., 1997 ▸) means that it is predominantly in the thiolate state (S−), making it highly susceptible to oxidation. The C215 Sγ atom forms a S–HN hydrogen bond with the G218 amide nitrogen [Fig. 1(C) ▸] (Mundlapati et al., 2015 ▸).
Oxidation of C215 to a cysteine sulfenic acid (C215-SOH) lengthens the hydrogen bond considerably [Fig. 1(C) ▸]. A proposed subsequent nucleophilic attack of the peptide NH of S216 on the electrophilic sulfur atom of Cys-SOH with concomitant release of water then leads to the formation of a sulfenyl-amide (C215-SN). The new C215-SN species fully disrupts the C215-G218 hydrogen bond, which opens the P-loop and is accompanied by major conformational changes in the nearby pTyr recognition loop [Fig. 1(D) ▸]. It has been proposed that reactive cysteines are enriched in strand-turn-helix motifs similar to those in PTP-1B, and that their strained cysteine backbone conformations position the adjacent residue to facilitate formation of a sulfenyl-amide (Defelipe et al., 2015 ▸). The various oxidation states of cysteine were discovered serendipitously in crystal structures of PTP-1B proteins exposed to several inhibitors (van Montfort et al., 2003 ▸). This suggests that evolutionary pressure imparted a protein micro-environment on these amino acids to promote high reactivity, underscoring the functional role of cysteine modification.
3. Cysteine modification alters the electrostatic and structural protein environment
In the large class of cysteine-dependent enzymes, the catalytic cysteine nucleophile is generally ionized to the thiolate anion, as discussed earlier. Because cysteine thiolates have a formal negative charge, their thiolate anions often accept charge-assisted hydrogen bonds, which stabilizes the thiolate and can increase the reactivity of the cysteine residue (Li et al., 2005 ▸; Roos et al., 2013 ▸). However, due to the Hammond postulate, the most reactive cysteine residues are those whose pK a is closely matched to ambient pH, and reaction rates then slow as the cysteine pK a values further decrease (Whitesides et al., 1977 ▸).
Covalent modification of the thiolate alters both its electrostatic and steric environment. Modification neutralizes the negative charge of the Sγ thiolate, making it a poorer hydrogen-bond acceptor [Fig. 1(C) ▸] and thereby reorganizing surrounding non-covalent interactions. Because cysteine-dependent enzymes employ covalent catalysis with a cysteine thiolate nucleophile, all such catalytic cysteine residues should experience a similar transient loss of negative charge. X-ray induced photooxidation of Cys-S− to Cys-SOH also results in thiolate charge neutralization. Therefore, radiation-driven Cys-SOH formation remodels the active site micro-environment in ways that are similar to on-pathway intermediates in many respects.
Remodeled hydrogen bond networks involving reactive cysteines can redistribute the protein conformational ensemble (Fig. 2 ▸), providing a mechanism for proteins to control conformational dynamics. Examining how these conformational changes propagate can connect molecular motions to function for proteins central to many cellular processes.
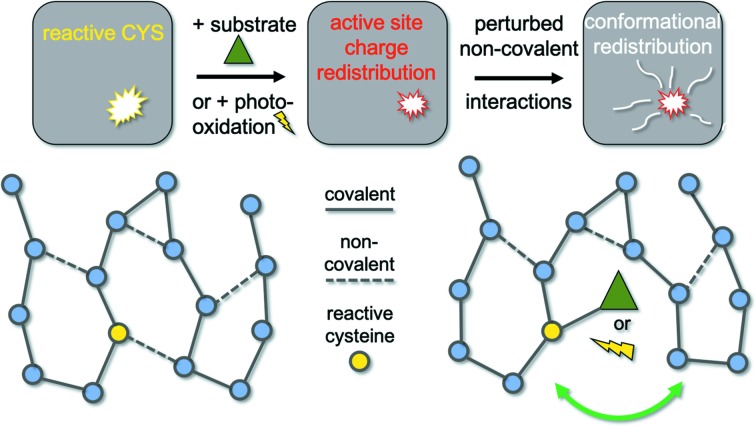
Figure 2.
Cysteine modification reorganizes non-covalent interaction networks similar to on-pathway catalytic intermediates. The graph networks depict a protein structure, with nodes representing residues, solid lines representing covalent bonds, and dashed lines representing non-covalent interactions. A reactive thiolate can be covalently modified through binding a substrate, or by a transient reaction intermediate. The covalent modification neutralizes the charge on the Sγ atom, changing the electrostatic microenvironment of the active site. X-ray induced photooxidation of Cys-S− to Cys-SOH equally results in thiolate charge neutralization. The modified charge of the Sγ acceptor atom considerably weakens or even disrupts any hydrogen bond. This altered non-covalent network can redistribute the protein conformational ensemble well beyond the active site microenvironment.
4. Cysteine modification is tunable by X-ray radiation and can report on functional motions
The physico-chemical origin of site-specific, X-ray induced cysteine photooxidation likely includes formation of ROS from nearby waters by irradiation and subsequent cysteine oxidation. This is similar to oxidation events arising from endogenously produced ROS in the cell, suggesting a roadmap for probing molecular mechanisms of cysteine-dependent enzymes using X-ray radiation. Our previous work established that proteins sample a restricted but functionally relevant conformational ensemble in a crystalline environment which is enriched at room temperature (Fraser et al., 2011 ▸). However, distinguishing functional motions from the many other equilibrium fluctuations of a macromolecule is challenging (Henzler-Wildman et al., 2007 ▸). Consequently, site-specific X-ray triggered structural changes (Schlichting et al., 2000 ▸; Colletier et al., 2008 ▸) combined with temperature-controlled crystallography (van den Bedem et al., 2013 ▸; Keedy, Kenner et al., 2015 ▸; Keedy, 2019 ▸) provide a powerful means to reveal structural mechanisms of macromolecular function for cysteine-dependent enzymes. Modification by X-ray radiation can provide a complementary approach to determine how the protein conformational ensemble redistributes in response to local, functionally important electrostatic and structural changes at cysteine residues. While care must be taken to minimize global radiation damage in data collection, the conformational ensemble at higher temperature is affected only mildly by global radiation damage, suggesting that the origin of enrichment is thermodynamic in nature (Russi et al., 2017 ▸) (Fig. 3 ▸). Site-specific radiation-induced changes should also be carefully monitored, as these are more sensitive to dose than global radiation damage processes (Bury & Garman, 2018 ▸).
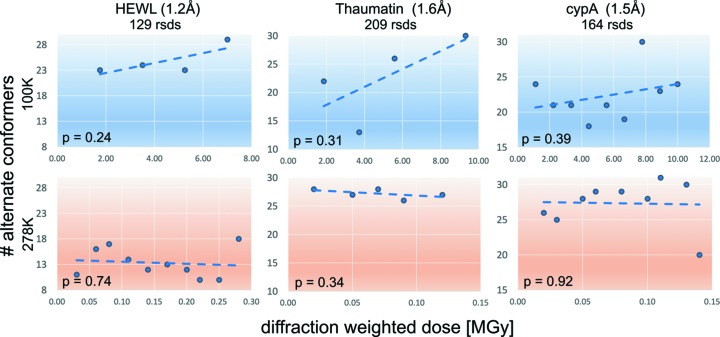
Figure 3.
The protein crystal conformational ensemble at ambient temperature is not dominated by global radiation damage [adapted from Russi et al. (2017 ▸)]. Analysis of conformational disorder with increasing radiation dose for three proteins. The slopes correspond to a linear regression of the number of distinct alternate conformers onto average diffraction weighted dose (Zeldin et al., 2013 ▸), with p-values reported at 95% confidence levels. The results did not identify a significant trend between dose and the observed protein conformational ensemble at cryogenic and room temperatures.
5. Interrogating conformational dynamics in ICH using site-specific cysteine photooxidation
The potential for uncovering molecular mechanisms in catalysis with site-specific radiation damage and temperature-controlled X-ray crystallography is exemplified by studies on isocyanide hydratase (ICH). ICH is a 230-residue, homodimeric cysteine-dependent enzyme of the DJ-1 superfamily that catalyzes the addition of water to diverse isocyanides to yield N-formamides (Lakshminarasimhan et al., 2010 ▸) [Fig. 4(A) ▸]. It is structurally similar to human DJ-1 (all non-hydrogen atom RMSD 1.5 Å), except for a long terminal helix that is absent in DJ-1. ICH is one of only two enzyme classes characterized to date that degrades isocyanides. Isocyanides frequently possess antimicrobial properties and are synthesized by microbes to kill competing species, explaining the prevalence of ICH homologs in fungi and bacteria that populate competitive microbial niches, particularly in the soil microbiome. Some isocyanides are validated antibiotics, including xanthocillin, darlucins A and B, and welwitindolinone (Scheuer, 1992 ▸).
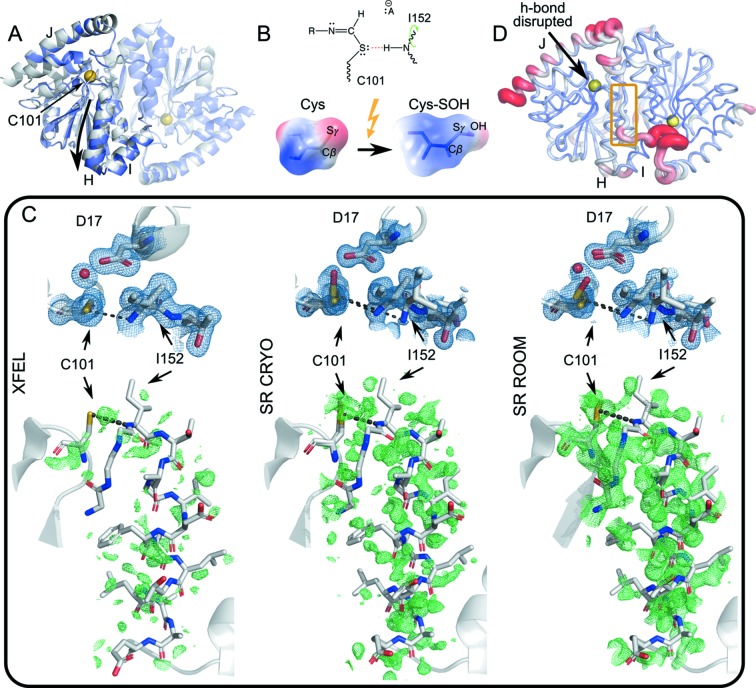
Figure 4.
Conformational dynamics during catalysis in ICH. (A) Homodimeric ICH. The ‘B’ protomer is shown transparent, and the catalytic cysteine Cys101 is indicated with a yellow sphere. The gray cartoon representation corresponds to the unshifted Cys101-S− enzyme. The slate representation is the shifted conformation corresponding to the Cys101-SOH state. While the entire dimer undergoes conformational redistribution, the shift is particularly pronounced for helix H (downward arrow) and linker IJ across the dimer interface. (B) The formation of a thioimidate intermediate neutralizes charge of the C101 Sγ atom (top diagram), weakening the Cys101-Sγ:Ile-NH hydrogen bond (red dashes). Photo-oxidation of Cys101-S− to Cys101-SOH has the same charge-neutralizing effect (bottom), illustrated by surfaces of electrostatic charge. (C) Top panels, 2mF o − DF c electron density maps (blue) of the active site contoured at 1σ. Bottom panels, mF o − DF c difference electron density maps (green) around helix H, phased with the XFEL conformer (PDB ID 6npq, resolution 1.6 Å), contoured at 2σ. The (unshifted) atom positions in the ‘SR-CRYO’ (PDB ID 6nja, resolution 1.05 Å) and ‘SR-ROOM’ (PDB ID 6ni6, resolution 1.2 Å) bottom panels are virtually identical to those of the XFEL conformer on the left. The conformational ensemble shifts as Cys101 is more fully oxidized, manifested as shifting occupancies of the Cys-S− and Cys-SOH states, from XFEL to synchrotron (SR) X-ray irradiation at room temperature. (D) ICH protein conformational dynamics most affected by the disruption of the Cys101-Sγ:Ile-NH hydrogen bond. We selected ten low free energy motion modes from a KFA analysis that overlapped least for the Cys-S− enzyme with the Sγ-NH hydrogen bond intact and without the Sγ-NH hydrogen bond in one of the dimer subunits. These motion modes are most affected by the change in hydrogen-bonding network. The figure shows the root-mean-square fluctuations when these motion modes are sampled. The IJ linker from the opposite dimer subunit appears strongly affected by the disruption (yellow box).
ICH catalyzes an uncommon reaction, capitalizing on the carbenic, electrophilic character of the isocyanide carbon atom to initiate attack at the catalytic cysteine residue [Fig. 4(B) ▸]. ICH is the first known example of such cysteine-dependent isocyanide chemistry in biochemistry, recapitulating features of the organic synthesis reactions (Ugi and Passerini reactions) that exploit the unusual electrophilic and nucleophilic character at isocyanide carbon atoms.
X-ray crystallographic studies of ICH showed photooxidation of the active-site cysteine residue (Cys101) to cysteine-sulfenic acid (Cys-SOH) in both cryocooled (100 K) (Lakshminarasimhan et al., 2010 ▸) and room-temperature (274–277 K) datasets (DasGupta et al., 2019 ▸). This is reminiscent of a similar, though less dramatic, cysteine photo-oxidation event observed for the homologous reactive cysteine residue in human DJ-1. In these examples, the local structural features that appear to be correlated with photo-oxidation of cysteine residues are a proximal ordered water molecule and a favorable hydrogen-bonding environment, both of which are commonly found in DJ-1 superfamily proteins. In ICH, Cys101 is located within 3 Å of an ordered water molecule that becomes the oxygen atom of the Cys101-SOH species, similar to the modification of Cys106 in DJ-1 [Fig. 4(C) ▸, top panels]. It is reasonable to speculate that this proximal water molecule is converted to HO during X-ray illumination, which then reacts with Cys101 in either the thiolate or thiyl radical form to generate Cys-SOH.
during X-ray illumination, which then reacts with Cys101 in either the thiolate or thiyl radical form to generate Cys-SOH.
We recently designed a series of radiation-dose-dependent perturbation experiments to monitor the effects of site-specific radiation damage at Cys101 in ICH, from minimal absorbed radiation dose at an XFEL to full site-specific oxidation using synchrotron radiation at ambient temperature (DasGupta et al., 2019 ▸) [Fig. 4(C) ▸, bottom panels]. qFit multi-conformer modeling, an automated and unbiased computational approach to identify alternative conformations and their associated occupancies in electron density maps (van den Bedem et al., 2009 ▸; Keedy, Fraser et al., 2015 ▸; van Zundert et al., 2018 ▸), of these data sets revealed a shifting conformational ensemble around the active site and the adjacent α-helix H as a consequence of changes in the active-site electrostatic environment owing to the formation of a cysteine sulfenic acid [Fig. 4(C) ▸].
6. Computational analyses can provide a mechanistic basis for shifting conformational ensembles
Functionally rationalizing conformational changes observed in crystallographic data sets without explicit information about the time evolution of events is not straightforward (van den Bedem & Fraser, 2015 ▸). To help connect the conformational shifts in ICH to the catalytic cycle we turned to molecular dynamics simulation. Spatiotemporal limitations on simulations have, until recently, hampered agreement with crystallographic experiments, but, largely in parallel to the resolution revolution in cryo-EM (Kühlbrandt, 2014 ▸), molecular dynamics simulation has experienced its own (spatiotemporal) resolution revolution (Dror et al., 2012 ▸). Inexpensive ‘prosumer’ GPUs now routinely enable microsecond time-scale simulations of protein crystals constituting multiple unit cells – improving agreement between simulation and several different types of experimental data (Janowski et al., 2016 ▸; Wall et al., 2014 ▸).
In ICH, we postulated that charge neutralization of Sγ owing to the formation of Cys-SOH weakened its role as an acceptor for an Sγ-NH hydrogen bond with the Ile152 amide nitrogen. Microsecond molecular dynamics simulations of Cys101 thiolate (–S−) and oxidized Cys-SOH protein crystals in 2 × 2 × 2 unit-cell simulation volumes confirmed that the Sγ-NH hydrogen bond disrupted readily under oxidized conditions, but remained largely intact in the thiolate state (DasGupta et al., 2019 ▸). Strikingly, the simulations revealed how conformational changes similar to those observed in the crystal structures centered on the active site, propagated through helix H and asymmetrically across the dimer through a highly flexible seven-residue linker between helices I and J of the opposite dimer subunit. The large conformational changes of helix H likely accommodate the later steps of catalysis, hydrolysis and product release.
Further analysis with kinematic flexibility analysis (KFA; Budday et al., 2018 ▸) affirmed that linker IJ is sensitive to changes in the hydrogen-bonding network, bridging an allosteric communication pathway between the ICH subunits and active sites [Fig. 4(D) ▸]. KFA is a computational method specifically designed to detect how protein motion modes adjust to changes in the non-covalent interaction network. KFA calculates protein orthogonal motion modes from treating hydrogen bonds and hydrophobic interactions as an explicit constraint network subject to relaxation by an energy penalty. It rank-orders motions by the magnitude of free energy changes incurred by non-covalent interactions and molecular rigidity (Budday et al., 2015 ▸). A similar, rigidity-based approach also provided a structural basis for allosteric communication between the active sites of fluoroacetate dehalogenase (Kim et al., 2017 ▸).
7. Biochemical validation of cysteine photo-oxidation effects is essential
Most examples of site-specific radiation damage involve a redox component. Therefore, radiation-driven modifications frequently occur in protein active sites where electron transfer is coupled to biochemical function. Because of the potential for cysteine modifications to be artifactual when using high-intensity synchrotron X-ray radiation sources, it is important to determine whether the structural and dynamic changes in proteins observed upon X-ray irradiation reflect physiologically relevant changes. In most cases, this validation requires identifying aspects of the radiation-driven modification that can be independently investigated using solution biochemistry methods in the absence of X-rays.
The proposed catalytic mechanism of ICH features a thioimidate intermediate formed at Cys101, which neutralizes the negative charge of Sγ [Fig. 4(B) ▸]. Protein dynamical events that are coupled to the charge state of cysteine (or other catalytic residues) should have detectable impact on enzyme turnover if they are functionally relevant. For example, carefully selected point mutations that modulate these dynamical aspects can validate structural hypotheses through comparison of steady-state and pre-steady-state kinetic behaviors with the wild-type protein. We emphasize that protein dynamics that are important for cysteine-dependent enzyme turnover are not necessarily important for transition state stabilization per se. Instead, they are most likely coupled to slower processes such as substrate binding, resolving intermediates, or product release. In many enzymes these steps are at least partially rate-limiting and thus amenable to kinetic characterization.
Redox modifications of cysteine residues are also potentially biochemically relevant. These include cysteine disulfides, direct oxidation to Cys-SOH, –SO2 − or SO3 −, or other, more exotic, modifications. Cysteine residues that are vulnerable to radiation damage in crystallo are often sensitive to modifications in the cell, as found for Cys106 in DJ-1 (Wilson et al., 2003 ▸). In these cases, it is imperative that biochemical validation of this cysteine modification be performed in cells using the growing variety of chemical probes and mass spectrometry approaches that monitor protein- and site-specific cysteine redox states (Reddie & Carroll, 2008 ▸). It should be borne in mind that the cysteine modification observed in the crystal may be different from that which is found in the cell, or that the modification is found only under a specific set of physiological conditions, such as oxidative stress, aging, or disease. Therefore, although a cautious approach should be taken when attempting to connect radiation-induced cysteine modification to physiological function, site-specific radiation damage of specific cysteine residues often suggests broader biological significance and thus justifies detailed biochemical follow up.
8. Conclusion
Reactive, functionally important cysteine residues can form radiation-induced sulfenic acids (Cys-SOH), which, like Asp and Glu decarboxylation, is an oxidative modification. In multiple proteins in the DJ-1 superfamily, photooxidation of cysteine appears to occur most readily when there is an ordered water molecule near (∼4 Å) the Sγ atom. We speculate that this nearby water molecule is converted to a hydroxyl radical ( OH), which subsequently oxidizes the nearby cysteine, although more complex redox chemistry resulting in the formation of H2O2 is also possible. Nevertheless, it is unclear why reactive cysteines would be favored targets of
OH), which subsequently oxidizes the nearby cysteine, although more complex redox chemistry resulting in the formation of H2O2 is also possible. Nevertheless, it is unclear why reactive cysteines would be favored targets of  OH modification, as this highly reactive radical ROS should be driving the reaction and thus would modify any nearby residue. Possible explanations include a contribution from thiyl radical formation as well, which may occur more readily for low pK
a cysteine residues, or that ordered water molecules are more likely to reside near reactive cysteine residues because thiolates are good hydrogen bond acceptors. Both hypotheses suggest that the low pK
a of a reactive cysteine thiol contributes to its susceptibility to photo-oxidation, which would correlate with reactivity in vivo. Perhaps due more to oversight than rarity, there are a limited number of proteins where such cysteine photo-oxidation is known to occur and searching for other validated examples would aid in establishing the structural determinants of cysteine photoreactivity.
OH modification, as this highly reactive radical ROS should be driving the reaction and thus would modify any nearby residue. Possible explanations include a contribution from thiyl radical formation as well, which may occur more readily for low pK
a cysteine residues, or that ordered water molecules are more likely to reside near reactive cysteine residues because thiolates are good hydrogen bond acceptors. Both hypotheses suggest that the low pK
a of a reactive cysteine thiol contributes to its susceptibility to photo-oxidation, which would correlate with reactivity in vivo. Perhaps due more to oversight than rarity, there are a limited number of proteins where such cysteine photo-oxidation is known to occur and searching for other validated examples would aid in establishing the structural determinants of cysteine photoreactivity.
Reactive cysteines typically have thiol pK a values that are several units lower than the unperturbed pK a of 8.3. Whether a cysteine is a thiol or thiolate in a crystal structure is determined by the difference in its pK a value and the pH of the buffer. Unfortunately, hydrogen atoms are difficult to observe directly with X-ray crystallography at typical resolutions and thus there is no reliable way of determining the ionization state of a cysteine residue from the electron density map alone. While it is common to add explicit hydrogen atoms to crystal structures using computational tools prior to deposition in the Protein Data Bank (PDB; Berman et al., 2000 ▸), the pK a values of atoms are only crudely considered. Cysteine residues are often modeled as protonated by default, which could lead to underreporting thiolates as hydrogen bond acceptors in the PDB. Indeed, a survey of the PDB revealed that the cysteine side-chain is fivefold more often reported as a hydrogen bond donor than as an acceptor (Mazmanian et al., 2016 ▸).
Cysteine modification is but one of the tools in the rapidly growing field of perturbation macromolecular crystallography (PMX) at XFELS and synchrotrons alike. In PMX, the protein crystal is subjected to a perturbation, for example laser excitation (Tenboer et al., 2014 ▸), mixing substrates (Kupitz et al., 2017 ▸), temperature (Keedy, Kenner et al., 2015 ▸), electric fields (Hekstra et al., 2016 ▸), etc., and its response is recorded. Technology development at lightsources, in X-ray detectors, and sample preparation have enabled this exciting approach to probe structural dynamics and conformational landscapes. Likewise, computational approaches like MD or QM/MM simulations and multi-conformer or ensemble modeling have matured sufficiently to generate and test hypotheses of crystallographic snapshots. However, independent biochemical experiments remain required to provide orthogonal validation.
Because all cysteine-dependent enzymes involve a covalent intermediate, we expect that many pharmaceutically interesting targets are amenable to interrogation by tuning photo-oxidation of the reactive cysteine. At short pulse lengths, XFEL light sources allow collection of damage-free reference structures and synchrotron radiation provides a dose- and energy-tunable X-ray source to fully characterize cysteine photoreactivity in diverse systems. However, care should be taken to question the physiological relevance of cysteine oxidation events until independently corroborated by multiple types of experiments. Many proteins rely on reactive cysteine residues for function, and thus there are abundant opportunities to expand our understanding of cysteine chemistry using X-rays as both probe and reactant.
Funding Statement
This work was funded by National Institute of General Medical Sciences grant GM123159 to Henry van den Bedem. Nebraska Tobacco Settlement Biomedical Research Development Fund grant .
References
- Aleyasin, H., Rousseaux, M. W. C., Phillips, M., Kim, R. H., Bland, R. J., Callaghan, S., Slack, R. S., During, M. J., Mak, T. W. & Park, D. S. (2007). Proc. Natl Acad. Sci. USA, 104, 18748–18753. [DOI] [PMC free article] [PubMed]
- Aron, L., Klein, P., Pham, T.-T., Kramer, E. R., Wurst, W. & Klein, R. (2010). PLoS Biol. 8, e1000349. [DOI] [PMC free article] [PubMed]
- Awoonor-Williams, E. & Rowley, C. N. (2018). J. Chem. Inf. Model. 58, 1935–1946. [DOI] [PubMed]
- Backus, K. M. (2019). Activity-Based Protein Profiling, edited by B. F. Cravatt, K.-L. Hsu & E. Weerapana, pp. 375–417. Cham: Springer International Publishing.
- Barbieri, L., Luchinat, E. & Banci, L. (2018). J. Biol. Inorg. Chem. 23, 61–69. [DOI] [PubMed]
- Bauer, R. A. (2015). Drug Discov. Today, 20, 1061–1073. [DOI] [PubMed]
- Bedem, H. van den, Bhabha, G., Yang, K., Wright, P. E. & Fraser, J. S. (2013). Nat. Methods, 10, 896–902. [DOI] [PMC free article] [PubMed]
- Bedem, H. van den, Dhanik, A., Latombe, J.-C. & Deacon, A. M. (2009). Acta Cryst. D65, 1107–1117. [DOI] [PMC free article] [PubMed]
- Bedem, H. van den & Fraser, J. S. (2015). Nat. Methods, 12, 307–318. [DOI] [PMC free article] [PubMed]
- Berman, H. M., Westbrook, J., Feng, Z., Gilliland, G., Bhat, T. N., Weissig, H., Shindyalov, I. N. & Bourne, P. E. (2000). Nucleic Acids Res. 28, 235–242. [DOI] [PMC free article] [PubMed]
- Blackinton, J., Lakshminarasimhan, M., Thomas, K. J., Ahmad, R., Greggio, E., Raza, A. S., Cookson, M. R. & Wilson, M. A. (2009). J. Biol. Chem. 284, 6476–6485. [DOI] [PMC free article] [PubMed]
- Böhmer, F., Szedlacsek, S., Tabernero, L., Östman, A. & den Hertog, J. (2013). FEBS J. 280, 413–431. [DOI] [PubMed]
- Bonifati, V., Rizzu, P., van Baren, M. J., Schaap, O., Breedveld, G. J., Krieger, E., Dekker, M. C. J., Squitieri, F., Ibanez, P., Joosse, M., van Dongen, J. W., Vanacore, N., van Swieten, J. C., Brice, A., Meco, G., van Duijn, C. M., Oostra, B. A. & Heutink, P. (2003). Science, 299, 256–259.
- Bonilha, V. L., Bell, B. A., Rayborn, M. E., Samuels, I. S., King, A., Hollyfield, J. G., Xie, C. & Cai, H. (2017). Free Radic. Biol. Med. 104, 226–237. [DOI] [PMC free article] [PubMed]
- Budday, D., Leyendecker, S. & van den Bedem, H. (2015). J. Mech. Phys. Solids, 83, 36–47. [DOI] [PMC free article] [PubMed]
- Budday, D., Leyendecker, S. & van den Bedem, H. (2018). J. Chem. Inf. Model. 58, 2108–2122. [DOI] [PMC free article] [PubMed]
- Bury, C. S. & Garman, E. F. (2018). J. Appl. Cryst. 51, 952–962.
- Canet-Avilés, R. M., Wilson, M. A., Miller, D. W., Ahmad, R., McLendon, C., Bandyopadhyay, S., Baptista, M. J., Ringe, D., Petsko, G. A. & Cookson, M. R. (2004). Proc. Natl Acad. Sci. USA, 101, 9103–9108. [DOI] [PMC free article] [PubMed]
- Cao, J., Lou, S., Ying, M. & Yang, B. (2015). Biochem. Pharmacol. 93, 241–250. [DOI] [PubMed]
- Cao, J., Ying, M., Xie, N., Lin, G., Dong, R., Zhang, J., Yan, H., Yang, X., He, Q. & Yang, B. (2014). Antioxid. Redox Signal. 21, 1443–1459. [DOI] [PMC free article] [PubMed]
- Casimiro-Garcia, A., Trujillo, J. I., Vajdos, F., Juba, B., Banker, M. E., Aulabaugh, A., Balbo, P., Bauman, J., Chrencik, J., Coe, J. W., Czerwinski, R., Dowty, M., Knafels, J. D., Kwon, S., Leung, L., Liang, S., Robinson, R. P., Telliez, J.-B., Unwalla, R., Yang, X. & Thorarensen, A. (2018). J. Med. Chem. 61, 10665–10699. [DOI] [PubMed]
- Colletier, J.-P., Bourgeois, D., Sanson, B., Fournier, D., Sussman, J. L., Silman, I. & Weik, M. (2008). Proc. Natl Acad. Sci. USA, 105, 11742–11747. [DOI] [PMC free article] [PubMed]
- DasGupta, M., Budday, D., Madzelan, P., Seravalli, J., Hayes, B., Sierra, R. G., Alonso-Mori, R., Brewster, A. S., Sauter, N. K., Applegate, G. A., Tiwari, V., Berkowitz, D. B., Thompson, M. C., Fraser, J. S., Wall, M. E., van den Bedem, H. & Wilson, M. A. (2019). bioRxiv 524751; doi: 10.1101/524751.
- Defelipe, L. A., Lanzarotti, E., Gauto, D., Marti, M. A. & Turjanski, A. G. (2015). PLoS Comput. Biol. 11, e1004051. [DOI] [PMC free article] [PubMed]
- Drechsel, J., Mandl, F. A. & Sieber, S. A. (2018). ACS Chem. Biol. 13, 2016–2019. [DOI] [PubMed]
- Dror, R. O., Dirks, R. M., Grossman, J. P., Xu, H. & Shaw, D. E. (2012). Annu. Rev. Biophys. 41, 429–452. [DOI] [PubMed]
- Fraser, J. S., van den Bedem, H., Samelson, A. J., Lang, P. T., Holton, J. M., Echols, N. & Alber, T. (2011). Proc. Natl Acad. Sci. USA, 108, 16247–16252. [DOI] [PMC free article] [PubMed]
- García-Santamarina, S., Boronat, S. & Hidalgo, E. (2014). Biochemistry, 53, 2560–2580. [DOI] [PubMed]
- Garman, E. F. & Weik, M. (2017). Protein Crystallography: Methods and Protocols, edited by A. Wlodawer, Z. Dauter & M. Jaskolski, pp. 467–489. New York: Springer.
- Gehringer, M. & Laufer, S. A. (2019). J. Med. Chem. doi:10.1021/acs.jmedchem.8b01153.
- George, G. N., Pickering, I. J., Pushie, M. J., Nienaber, K., Hackett, M. J., Ascone, I., Hedman, B., Hodgson, K. O., Aitken, J. B., Levina, A., Glover, C. & Lay, P. A. (2012). J. Synchrotron Rad. 19, 875–886. [DOI] [PMC free article] [PubMed]
- Guzman, J. N., Sanchez-Padilla, J., Wokosin, D., Kondapalli, J., Ilijic, E., Schumacker, P. T. & Surmeier, D. J. (2010). Nature, 468, 696–700. [DOI] [PMC free article] [PubMed]
- Hekstra, D. R., White, K. I., Socolich, M. A., Henning, R. W., Šrajer, V. & Ranganathan, R. (2016). Nature, 540, 400–405. [DOI] [PMC free article] [PubMed]
- Henzler-Wildman, K. A., Thai, V., Lei, M., Ott, M., Wolf-Watz, M., Fenn, T., Pozharski, E., Wilson, M. A., Petsko, G. A., Karplus, M., Hübner, C. G. & Kern, D. (2007). Nature, 450, 838–844. [DOI] [PubMed]
- Janowski, P. A., Liu, C., Deckman, J. & Case, D. A. (2016). Protein Sci. 25, 87–102. [DOI] [PMC free article] [PubMed]
- Joselin, A. P., Hewitt, S. J., Callaghan, S. M., Kim, R. H., Chung, Y.-H., Mak, T. W., Shen, J., Slack, R. S. & Park, D. S. (2012). Hum. Mol. Genet. 21, 4888–4903. [DOI] [PubMed]
- Julien, S. G., Dubé, N., Hardy, S. & Tremblay, M. L. (2011). Nat. Rev. Cancer, 11, 35–49. [DOI] [PubMed]
- Keedy, D. A. (2019). Acta Cryst. D75, 123–137. [DOI] [PMC free article] [PubMed]
- Keedy, D. A., Fraser, J. S. & van den Bedem, H. (2015). PLoS Comput. Biol. 11, e1004507. [DOI] [PMC free article] [PubMed]
- Keedy, D. A., Kenner, L. R., Warkentin, M., Woldeyes, R. A., Hopkins, J. B., Thompson, M. C., Brewster, A. S., Van Benschoten, A. H., Baxter, E. L., Uervirojnangkoorn, M., McPhillips, S. E., Song, J., Alonso-Mori, R., Holton, J. M., Weis, W. I., Brunger, A. T., Soltis, S. M., Lemke, H., Gonzalez, A., Sauter, N. K., Cohen, A. E., van den Bedem, H., Thorne, R. E. & Fraser, J. S. (2015). eLife, 4, e07574. [DOI] [PMC free article] [PubMed]
- Kim, T. H., Mehrabi, P., Ren, Z., Sljoka, A., Ing, C., Bezginov, A., Ye, L., Pomès, R., Prosser, R. S. & Pai, E. F. (2017). Science, 355, eaag2355. [DOI] [PubMed]
- Kühlbrandt, W. (2014). Science, 343, 1443–1444. [DOI] [PubMed]
- Kupitz, C., Olmos, J. L. Jr, Holl, M., Tremblay, L., Pande, K., Pandey, S., Oberthür, D., Hunter, M., Liang, M., Aquila, A., Tenboer, J., Calvey, G., Katz, A., Chen, Y., Wiedorn, M. O., Knoska, J., Meents, A., Majriani, V., Norwood, T., Poudyal, I., Grant, T., Miller, M. D., Xu, W., Tolstikova, A., Morgan, A., Metz, M., Martin-Garcia, J. M., Zook, J. D., Roy-Chowdhury, S., Coe, J., Nagaratnam, N., Meza, D., Fromme, R., Basu, S., Frank, M., White, T., Barty, A., Bajt, S., Yefanov, O., Chapman, H. N., Zatsepin, N., Nelson, G., Weierstall, U., Spence, J., Schwander, P., Pollack, L., Fromme, P., Ourmazd, A., Phillips, G. N. Jr & Schmidt, M. (2017). Struct. Dyn. 4, 044003. [DOI] [PMC free article] [PubMed]
- Lakshminarasimhan, M., Madzelan, P., Nan, R., Milkovic, N. M. & Wilson, M. A. (2010). J. Biol. Chem. 285, 29651–29661. [DOI] [PMC free article] [PubMed]
- Lee, J.-Y., Song, J., Kwon, K., Jang, S., Kim, C., Baek, K., Kim, J. & Park, C. (2012). Hum. Mol. Genet. 21, 3215–3225. [DOI] [PubMed]
- Li, H., Robertson, A. D. & Jensen, J. H. (2005). Proteins, 61, 704–721. [DOI] [PubMed]
- Liu, Q., Sabnis, Y., Zhao, Z., Zhang, T., Buhrlage, S. J., Jones, L. H. & Gray, N. S. (2013). Chem. Biol. 20, 146–159. [DOI] [PMC free article] [PubMed]
- Lohse, D. L., Denu, J. M., Santoro, N. & Dixon, J. E. (1997). Biochemistry, 36, 4568–4575. [DOI] [PubMed]
- Maleknia, S. D., Brenowitz, M. & Chance, M. R. (1999). Anal. Chem. 71, 3965–3973. [DOI] [PubMed]
- Marinho, H. S., Real, C., Cyrne, L., Soares, H. & Antunes, F. (2014). Redox Biol. 2, 535–562. [DOI] [PMC free article] [PubMed]
- Mazmanian, K., Sargsyan, K., Grauffel, C., Dudev, T. & Lim, C. (2016). J. Phys. Chem. B, 120, 10288–10296. [DOI] [PubMed]
- Montfort, R. L. M. van, Congreve, M., Tisi, D., Carr, R. & Jhoti, H. (2003). Nature, 423, 773–777. [DOI] [PubMed]
- Mullett, S. J. & Hinkle, D. A. (2011). J. Neurochem. 117, 375–387. [DOI] [PMC free article] [PubMed]
- Mundlapati, V. R., Ghosh, S., Bhattacherjee, A., Tiwari, P. & Biswal, H. S. (2015). J. Phys. Chem. Lett. 6, 1385–1389. [DOI] [PubMed]
- Nagakubo, D., Taira, T., Kitaura, H., Ikeda, M., Tamai, K., Iguchi-Ariga, S. M. & Ariga, H. (1997). Biochem. Biophys. Res. Commun. 231, 509–513. [DOI] [PubMed]
- Ostrem, J. M., Peters, U., Sos, M. L., Wells, J. A. & Shokat, K. M. (2013). Nature, 503, 548–551. [DOI] [PMC free article] [PubMed]
- Owen, R. L., Axford, D., Nettleship, J. E., Owens, R. J., Robinson, J. I., Morgan, A. W., Doré, A. S., Lebon, G., Tate, C. G., Fry, E. E., Ren, J., Stuart, D. I. & Evans, G. (2012). Acta Cryst. D68, 810–818. [DOI] [PMC free article] [PubMed]
- Peralta, D., Bronowska, A. K., Morgan, B., Dóka, É., Van Laer, K., Nagy, P., Gräter, F. & Dick, T. P. (2015). Nat. Chem. Biol. 11, 156–163. [DOI] [PubMed]
- Reddie, K. G. & Carroll, K. S. (2008). Curr. Opin. Chem. Biol. 12, 746–754. [DOI] [PubMed]
- Richarme, G., Mihoub, M., Dairou, J., Bui, L. C., Leger, T. & Lamouri, A. (2015). J. Biol. Chem. 290, 1885–1897. [DOI] [PMC free article] [PubMed]
- Roos, G., Foloppe, N. & Messens, J. (2013). Antioxid. Redox Signal. 18, 94–127. [DOI] [PubMed]
- Russi, S., González, A., Kenner, L. R., Keedy, D. A., Fraser, J. S. & van den Bedem, H. (2017). J. Synchrotron Rad. 24, 73–82. [DOI] [PMC free article] [PubMed]
- Saito, Y., Miyasaka, T., Hatsuta, H., Takahashi-Niki, K., Hayashi, K., Mita, Y., Kusano-Arai, O., Iwanari, H., Ariga, H., Hamakubo, T., Yoshida, Y., Niki, E., Murayama, S., Ihara, Y. & Noguchi, N. (2014). J. Neuropathol. Exp. Neurol. 73, 714–728. [DOI] [PMC free article] [PubMed]
- Salmeen, A., Andersen, J. N., Myers, M. P., Meng, T.-C., Hinks, J. A., Tonks, N. K. & Barford, D. (2003). Nature, 423, 769–773. [DOI] [PubMed]
- Scheuer, P. J. (1992). Acc. Chem. Res. 25, 433–439.
- Schlichting, I., Berendzen, J., Chu, K., Stock, A. M., Maves, S. A., Benson, D. E., Sweet, R. M., Ringe, D., Petsko, G. A. & Sligar, S. G. (2000). Science, 287, 1615–1622. [DOI] [PubMed]
- Tashiro, S., Caaveiro, J. M. M., Nakakido, M., Tanabe, A., Nagatoishi, S., Tamura, Y., Matsuda, N., Liu, D., Hoang, Q. Q. & Tsumoto, K. (2018). ACS Chem. Biol. 13, 2783–2793. [DOI] [PMC free article] [PubMed]
- Tashiro, S., Caaveiro, J. M. M., Wu, C.-X., Hoang, Q. Q. & Tsumoto, K. (2014). Biochemistry, 53, 2218–2220. [DOI] [PubMed]
- Tenboer, J., Basu, S., Zatsepin, N., Pande, K., Milathianaki, D., Frank, M., Hunter, M., Boutet, S., Williams, G. J., Koglin, J. E., Oberthuer, D., Heymann, M., Kupitz, C., Conrad, C., Coe, J., Roy-Chowdhury, S. U., Weierstall, U., James, D., Wang, D., Grant, T., Barty, A., Yefanov, O., Scales, J., Gati, C., Seuring, C., Srajer, V., Henning, R., Schwander, P., Fromme, R., Ourmazd, A., Moffat, K., Van Thor, J. J., Spence, J. C. H., Fromme, P., Chapman, H. N. & Schmidt, M. (2014). Science, 346, 1242–1246. [DOI] [PMC free article] [PubMed]
- Truong, T. H., Ung, P. M.-U., Palde, P. B., Paulsen, C. E., Schlessinger, A. & Carroll, K. S. (2016). Cell. Chem. Biol. 23, 837–848. [DOI] [PMC free article] [PubMed]
- Visscher, M., Arkin, M. R. & Dansen, T. B. (2016). Curr. Opin. Chem. Biol. 30, 61–67. [DOI] [PMC free article] [PubMed]
- Wall, M. E., Van Benschoten, A. H., Sauter, N. K., Adams, P. D., Fraser, J. S. & Terwilliger, T. C. (2014). Proc. Natl Acad. Sci. USA, 111, 17887–17892. [DOI] [PMC free article] [PubMed]
- Wang, J. (2016). Protein Sci. 25, 1407–1419. [DOI] [PMC free article] [PubMed]
- Whitesides, G. M., Lilburn, J. E. & Szajewski, R. P. (1977). J. Org. Chem. 42, 332–338.
- Wilson, M. A., Collins, J. L., Hod, Y., Ringe, D. & Petsko, G. A. (2003). Proc. Natl Acad. Sci. USA, 100, 9256–9261. [DOI] [PMC free article] [PubMed]
- Winterbourn, C. C. (2008). Nat. Chem. Biol. 4, 278–286. [DOI] [PubMed]
- Witt, A. C., Lakshminarasimhan, M., Remington, B. C., Hasim, S., Pozharski, E. & Wilson, M. A. (2008). Biochemistry, 47, 7430–7440. [DOI] [PMC free article] [PubMed]
- Xu, G. & Chance, M. R. (2005). Anal. Chem. 77, 2437–2449. [DOI] [PubMed]
- Xu, S., Yang, X., Qian, Y. & Xiao, Q. (2018). J. Neurochem. 145, 312–322. [DOI] [PubMed]
- Yip, S.-C., Saha, S. & Chernoff, J. (2010). Trends Biochem. Sci. 35, 442–449. [DOI] [PMC free article] [PubMed]
- Young, D., Pedre, B., Ezeriņa, D., De Smet, B., Lewandowska, A., Tossounian, M.-A., Bodra, N., Huang, J., Astolfi Rosado, L., Van Breusegem, F. & Messens, J. (2019). Antioxid. Redox Signal. 30, 1285–1324. [DOI] [PubMed]
- Zeldin, O. B., Brockhauser, S., Bremridge, J., Holton, J. M. & Garman, E. F. (2013). Proc. Natl Acad. Sci. USA, 110, 20551–20556. [DOI] [PMC free article] [PubMed]
- Zundert, G. C. P. van, Hudson, B. M., de Oliveira, S., Keedy, D. A., Fonseca, R., Heliou, A., Suresh, P., Borrelli, K., Day, T., Fraser, J. & van den Bedem, H. (2018). J. Med. Chem. 61, 11183–11198. [DOI] [PMC free article] [PubMed]