Fig. 4.
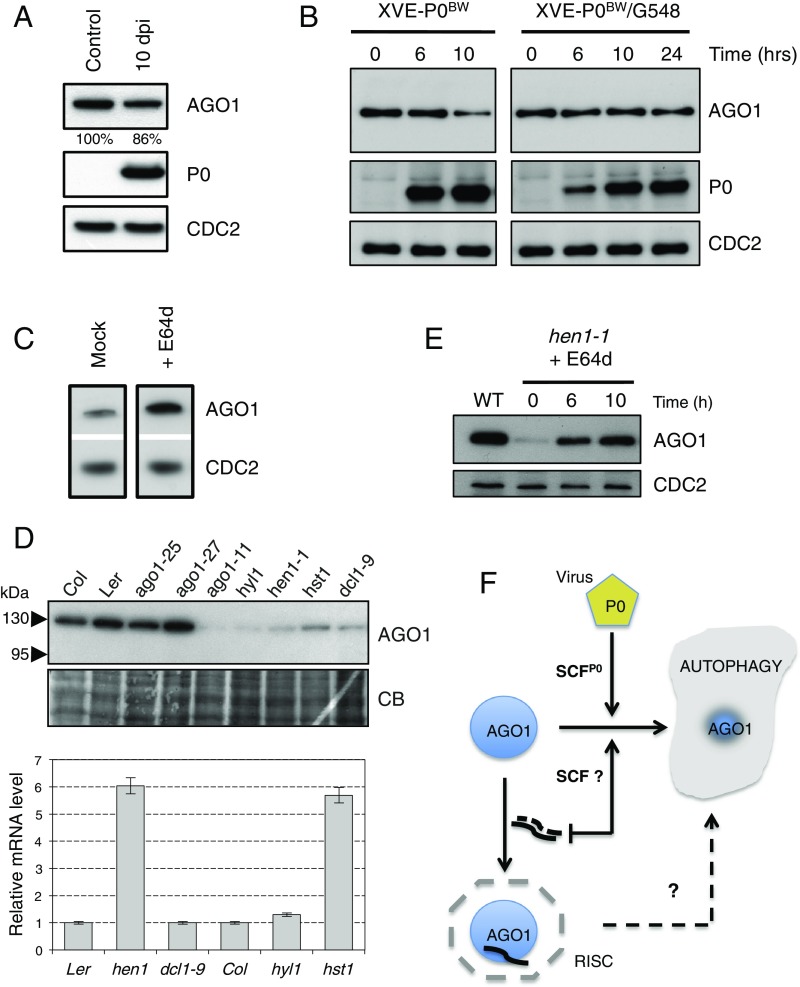
P0-mediated degradation of AGO1 is compromised in amsh3-1 mutant and in TOR-overexpressing plants and the endogenous pathway for AGO1 degradation also relies on autophagy. (A) Comparison of AGO1 protein accumulation level between uninduced 10-d–old XVE-P0BW-myc/amsh3-1 seedlings (control) and P0-induced 10-d-old XVE-P0BW-myc/amsh3-1 seedlings. For P0 induction, plants have been germinated and grown for 10 d on MS-agar dishes supplemented with 10 μM β-estradiol [10 d passed on induction (dpi)]. (B) AGO1 degradation kinetics performed on 7-d old XVE-P0BW-myc and XVE-P0BW-myc/G548 seedlings treated with β-estradiol (5 μM) for P0-myc induction. Because P0 induction is delayed in the XVE-P0BW-myc/G548 line, we extended this kinetic to 24 h. AGO1, P0-myc, and CDC2 (loading control) protein contents were assayed by Western blot. (C) E64d treatment (20 μM for 24 h) on wild-type Col-0 seedling leads to a higher accumulation of AGO1 protein. (D) Mutants affected in miRNA maturation and production pathways show reduced level of AGO1. (Upper) AGO1 protein accumulation in mutants and wild-type controls assayed by Western blot using the anti-AGO1 antibody. Coomassie blue staining is given as loading control. (Lower) AGO1 mRNA accumulation in each mutant assayed by quantitative RT-PCR. (E) The hen1-1 seedlings treated with E64d (20 μM) show AGO1 protein reaccumulation. AGO1 and CDC2 (loading control) protein contents were assayed by Western blot at the indicated time point. (F) Model for AGO1 turnover in a viral and nonviral context.